Abstract
Simian Immunodeficiency Virus (SIV), like its human homologues (HIV-1, HIV-2), requires a −1 translational frameshift event to properly synthesize all of the proteins required for viral replication. The frameshift mechanism is dependent upon a seven nucleotide slippery sequence and a downstream RNA structure. In SIV, the downstream RNA structure has been proposed to be either a stem-loop or a pseudoknot. Here, we report the functional, structural and thermodynamic characterization of the SIV frameshift site RNA. Translational frameshift assays indicate that a stem-loop structure is sufficient to promote efficient frameshifting in vitro. NMR and thermodynamic studies of SIV RNA constructs of varying length further support the absence of any pseudoknot interaction and indicate the presence of a stable stem-loop structure. We determined the structure of the SIV frameshift-inducing RNA by NMR. The structure reveals a highly ordered 12 nucleotide loop containing a sheared G-A pair, cross-strand adenine stacking, two G-C base-pairs, and a novel CCC triloop turn. The loop structure and its high thermostability preclude pseudoknot formation. Sequence conservation and modeling studies suggest that HIV-2 RNA forms the same structure. We conclude that, like the main sub-groups of HIV-1, SIV and HIV-2 utilize stable stem-loop structures to function as a thermodynamic barrier to translation, thereby inducing ribosomal pausing and frameshifting.
Introduction
Simian immunodeficiency virus (SIV), like human immunodeficiency virus (HIV), requires a programmed −1 translational frameshift between the gag and pol coding regions of the viral mRNA.1 The −1 frameshift allows for stop codon read-through at the end of the gag gene, and continued synthesis of the protein encoded by pol. This results in production of a Gag-Pol polyprotein fusion product, which upon proteolysis in immature virus particles will result in the protease, reverse transcriptase, and integrase enzymes, as well as the matrix, capsid and nucleocapsid structural proteins. These structural and enzymatic proteins are found in approximately a 10:1 to 20:1 molar ratio in HIV type 1 (HIV-1), depending on the assay used.2-8 This specific stoichiometry is required for appropriate packaging and formation of virus particles.8-10
In retroviruses, the −1 translational frameshift is programmed by two cis-acting RNA elements: a seven nucleotide slippery sequence, and a downstream RNA structure.11 In SIV, as in HIV-1,2 the slippery sequence is UUUUUUA, and is universally conserved among all isolates analyzed (Los Alamos National Laboratories HIV Database, http://www.hiv.lanl.gov). In this work, we focus on a subtype of SIV (SIVSMM) found in sooty mangabys (Cercocebus torquatus atys) and macaques (genus Macaca). This subtype is closely related to HIV type 2 (HIV-2),12-14 while SIV from chimpanzees (Pan troglodytes) is more closely related to HIV-1.12-15
Two models have been proposed for the SIVSMM downstream RNA secondary structure, a stem-loop,2 and an H-type pseudoknot16,17 (Figure 1). The pseudoknot model is based on the fact that seven nucleotides in the loop are complimentary to a downstream sequence; however, due to the high degree of sequence conservation, there is no observable covariance between these sequences that could be used to help validate the pseudoknot model. No structural data exists for the SIV frameshift-inducing RNA.
Figure 1.
Two secondary structure models for the SIV RNA frameshift-inducing element. The seven nucleotide slippery sequence is underlined in each model. (A) Previously proposed stem-loop model.2 (B) Proposed pseudoknot model.16,17 (C-E). RNA constructs studied by NMR. (C). SIV14−70, encompassing nucleotides 14−70 downstream of the slippery site. (D). SIV17−70. The first two C-G base-pairs were flipped to G-C pairs to facilitate in vitro transcription, and are shown in outline. Note that no base pairs are indicated within the pseudoknot portions of the secondary structures in (C) and (D), since the pseudoknot interaction has not been proven and is one of the subjects of this study. (E). SIV17−50. The first two base-pairs are as in (D).
In this study, our aims were to: 1) Determine the sequence requirement for frameshifting in SIVSMM; 2) Determine the structure of the SIVSMM frameshift-inducing RNA; 3) Analyze the thermodynamic properties of the structure, and 4) Compare the SIVSMM structure to HIV-118-20 and HIV-2 to determine if any structural conservation exists.
Results
SIV sequence-induced frameshifting in vitro
The sequence requirement for SIVSMM programmed translational frameshifting was investigated using a dual luciferase reporter assay.21,22 Briefly, SIVSMM sequences of varying length were cloned between Renilla and firefly luciferase reporter genes. In vitro translation of the frameshift reporter constructs was performed in rabbit reticulocyte lysate. Frameshift efficiencies were calculated by comparing the experimental and control levels of firefly luciferase activity, after normalizing their Renilla luciferase activities as previously described.21,22
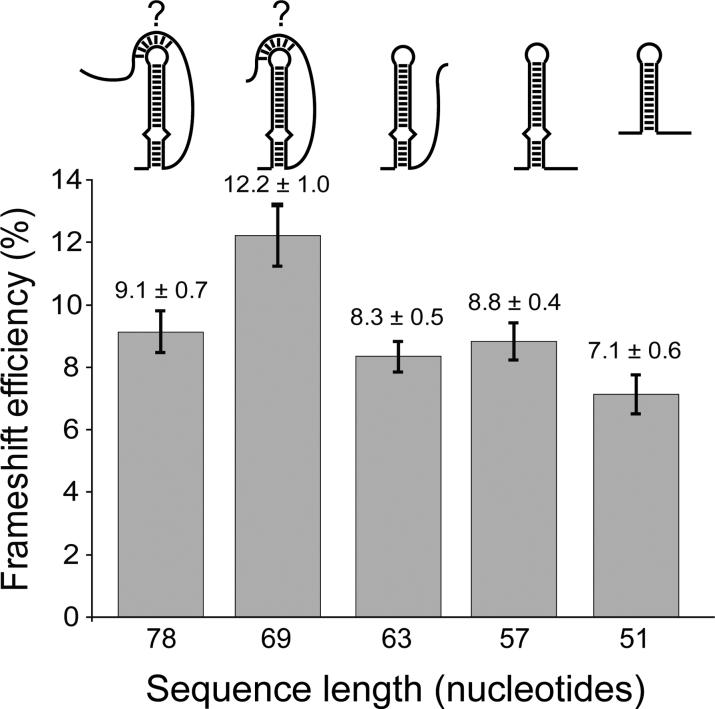
Results for all sequences tested showed frameshifting efficiencies from 7 to 12% (Figure 2). These frameshifting frequencies are very similar to those measured for HIV-1 in vitro.2-8 A 78-nucleotide sequence comprising all viral sequences necessary to form the potential pseudoknot (Figure 1B) induced frameshifting to a level of 9.1 ± 0.7 % (Figure 2). There is no significant difference between this value and the frameshifting frequencies observed for shorter constructs that cannot form a pseudoknot (e.g. 57 nt, 8.8 ± 0.4 %). Sequences of 69, 63, 57, and 51-nucleotides induced frameshifting to 12.2%, 8.3%, 8.8%, and 7.1%, respectively (Figure 2). These values were determined from 10 replicate measurements obtained from the same preparations of RNA and lysate. However, higher variability (+/− 3%) in frameshifting is observed when comparing different preparations of lysate and RNA (data not shown). Regardless of the relatively small differences between the various constructs, the data indicate that pseudoknot formation is not a requirement for programmed translational frameshifting by SIV, since the 63, 57 and 51-nucleotide constructs cannot form pseudoknots but produce frameshifting efficiencies comparable to the 78-nucleotide construct. The increase in frameshift efficiency for the 69-nucleotide sequence may be due to the variability of the assay system, or it may possibly be more optimally folded in the context of the reporter gene.
Figure 2.
Results of the in vitro frameshifting assay using dual-luciferase reporters.21 The length of the SIV sequences analyzed is indicated and compared to the frameshifting efficiency, as determined by an average of at 10 independent experiments, with the average value and standard error mean indicated above each column.
NMR Analysis of RNA folding
The stem-loop and pseudoknot models for the SIVSMM RNA were further investigated by NMR using three RNA constructs: 1. A 57 nucleotide sequence spanning nucleotides 14−70 (SIV14−70), which includes all nucleotides downstream of the slippery site involved in the proposed pseudoknot (Figure 1C). 2. A 54 nucleotide sequence that encompasses nucleotides 17−70 (SIV17−70) (Figure 1D). 3. A 34 nucleotide sequence that spans nucleotides 17−50 (SIV17−50) that can only form a stem-loop structure (Figure 1E). For constructs SIV17−70 and SIV17−50, the tandem C-G pairs at positions 17 and 18 where inverted to G-C pairs to facilitate transcription. Inversion of these C-G pairs had no affect on the folding of the RNA (vide infra, and Figure 3).
Figure 3.
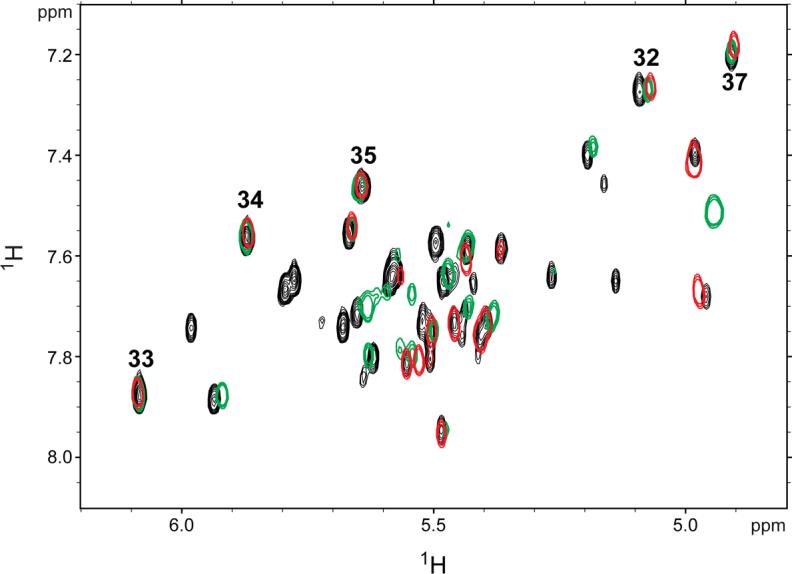
Overlay of 1H-1H TOCSY spectra obtained for SIV17−50 (red) and SIV17−70 (black) and SIV14−70 (green) at 750 MHz and 35°C in 10 mM sodium phosphate buffer (pH 7.0). Peaks corresponding to the loop cytidine peaks are numbered (32−35, 37).
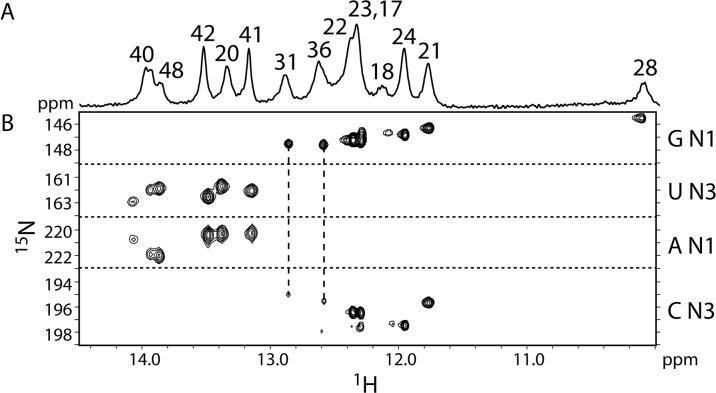
The chemical shifts in the RNA loop and proximal stem are essentially identical for all constructs (Figure 3). In particular, the four contiguous cytidines at the apex of the loop (C32−35), which would be involved in the potential pseudoknot interaction (if present), overlay well. There are no changes in chemical shifts for C33−35 and C37 in the various constructs. Nucleotide C32 exhibits very minor chemical shift changes, but these are likely due to differences between sample preparations, since the chemical shifts for this nucleotide vary slightly even between different preparations of the same RNA sequence (data not shown). Other differences in the NMR spectra of the various constructs are solely due to the different lengths and helical sequences of the various constructs. Additionally, 2D NOESY experiments reveal the presence of the same NOEs in the loop for all constructs (data not shown). All of the imino protons and their hydrogen-bond acceptors involved in Watson-Crick base pairing have been assigned for SIV17−50 (Figure 1E), which cannot form a pseudoknot (Figure 4). The same imino proton chemical shifts are observed for the two longer constructs (Figure 1C and D), and upon addition of 10 mM magnesium, no new imino peaks indicative of new Watson-Crick base pairs are observed (data not shown). These results indicate that the RNA adopts the same stem-loop structure in all constructs regardless of ionic conditions. Therefore, the in vitro translation data (Figure 2) and the NMR data (Figure 3) suggest that the SIVSMM RNA structure is a stem-loop, and not a pseudoknot.
Figure 4.
Direct detection of base-pairs in the SIV17−50 frameshift-inducing stem-loop. (A) 750 MHz 1D 1H NMR spectrum of the imino protons. Assignments are indicated by numbers above the peaks. (B) 800 MHz 2D HNN-COSY spectrum for direct detection of trans-hydrogen-bond scalar couplings. Two G-C pairs in the loop are indicated by dashed lines.
Structure and thermodynamics of the SIV RNA stem-loop
We further investigated the structure of the SIV17−50 RNA by NMR. Thirteen Watson-Crick base-pairs were confirmed by direct detection of trans-hydrogen-bond scalar couplings by HNN-COSY (Figure 4). Eleven of the Watson-Crick base-pairs correspond to the stem, while two additional G-C pairs were observed to form across the 12 nucleotide loop, between nucleotides G31-C37 and C32-G36. These assignments were confirmed by thermodynamic measurements and 1D NMR analysis of constructs with mutations of these loop nucleotides. Two cooperative melting transitions are observed: a lower-temperature transition at 66°C and a higher-temperature transition at 84°C (Figure 5A). These transitions can be assigned to the loop and stem regions of the molecule, respectively, by reference to experiments conducted on sequences that contain guanosine-to-cytidine mutations in the loop (Figure 5B), and also 1D NMR measurements as a function of temperature (data not shown). These data indicate that the stem is very stable, and the loop has a surprising degree of stability for a structure which is penalized 7.1 kcal/mole by mfold secondary structure prediction software.23 The 66°C melting transition for the loop is also observed for the longer SIV14−70 construct (Figure 1C and Supplemental data).
Figure 5.
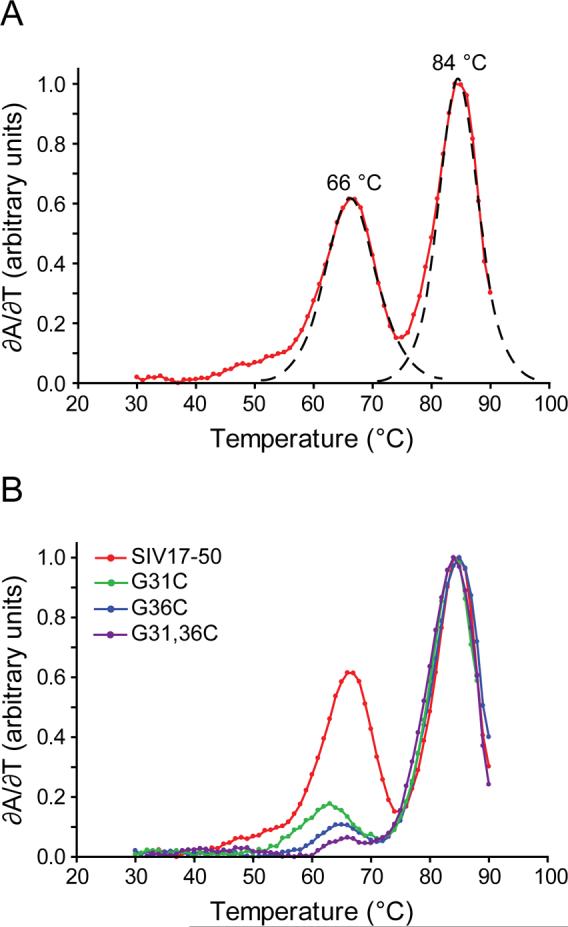
RNA thermal denaturation profiles for SIV17−50. (A) First derivative plot of change in UV absorbance vs. temperature (red), with fitted curves showing the deconvoluted transitions as dashed lines. The Tm values are shown above each peak. (B) First derivative plot of change in UV absorbance vs. temperature of SIV17−50 (red) compared to loop sequence mutants G31C (green), G36C (blue) and G31C, G36C (magenta).
The hydrogen-bonding data (Figure 4) and thermodynamic measurements (Figure 5) provide evidence for a highly structured loop conformation. NOESY data also provide further evidence for an unusually structured loop. A strong intranucleotide H6-H1' NOE and a relatively weak intranucleotide H6-H2' NOE for C35 indicate that the base predominately adopts the syn conformation (Supplemental data).24 Additionally, cross-strand internucleotide NOEs are observed between A29 and A39 (data not shown).
Structures of the SIV frameshift-inducing stem-loop were calculated using 647 NOE-derived distance restraints, backbone torsion angle restraints for the helical regions, hydrogen-bond restraints derived from scalar couplings, and 28 RDC restraints (Figure 4 and Table 1). The 20 lowest energy structures superimpose over all heavy atoms to an r.m.s.d. of 1.29 Å (Figure 6A and Table 1). The loop consists of a “neck” region (Figure 6, red nucleotides), and a “head” region (Figure 6, green nucleotides). The neck region is comprised of a sheared G-A pair (G28-A39) followed by a cross-strand adenine stack (A29 on A39), with A30 flipped out into the minor groove. The head region is comprised of two G-C pairs across the loop (G31-C37 and C32-G36) which close a compact three cytidine triloop (C33,C34,C35). The 12-nucleotide loop structure is well defined, and converges to an r.m.s.d. of 1.13 Å (Figure 6B and Table 1), though the A-form helical stem is more precisely defined as is usually the case for RNA solution structures (Figure 6C and Table 1).
Table 1.
| NOE-derived distance restraints | 647 |
| Intranucleotide | 253 |
| Internucleotide | 394 |
| Dihedral restraints | 219 |
| Hydrogen-bond restraints | 36 |
| RDC restraints | 28 |
| r.m.s.d. (all heavy atoms) | 1.29 Å |
| over stem (residues 17−27, 40−50) | 0.61 Å |
| over loop (residues 28−39) | 1.13 Å |
| Average NOE r.m.s.d. (Å) | 0.018 |
| Average dihedral r.m.s.d. (o) | 0.40 |
| Average RDC r.m.s.d. (Hz) | 1.46 |
Figure 6.

NMR structures of the SIV17−50 frameshift-inducing RNA stem-loop. (A) The 20 lowest energy structures superimposed over all heavy atoms. The A-form stem is shown in blue, the purine loop nucleotides 28−30 and 38−39 are shown are in red, and the remaining loop nucleotides 31−37 are shown in green. (B) Structures superimposed over all nucleotides in the 12-nucleotide loop (G28-A39). (C) Structures superimposed over all nucleotides in the stem (nucleotides 17−27 and 40−50). (D) Stereo view of the lowest energy structure.
Discussion
In this study, we have shown that the −1 translational frameshift in SIV is programmed by a thermodynamically stable RNA stem-loop. Analysis of frameshift efficiencies of various constructs suggests that pseudoknot formation does not play a significant role in translational frameshifting, at least in vitro. Furthermore, we observe that the SIV loop is structured in a manner that precludes pseudoknot formation, since two of the nucleotides that would participate in such a pseudoknot are sequestered within stable Watson-Crick base-pairings across the loop. Hence, the 12 nucleotide loop is reduced to a compact CCC triloop conformation. To the best of our knowledge, the CCC triloop is a novel turn conformation. The triloop conformation consists of a base-stack of C33 on C32, with C34 flipped into the minor groove, and C35 adopting a syn-conformation (Figure 7A). The syn conformation of C35 is consistent with the observed intensity of the NOE between its H1' and H6 protons (Supplemental Data). The unique CCC triloop structure may be stabilized by a potential hydrogen-bond between the carboxyl oxygen of C34 and the ribose 2'OH of C37.
Figure 7.
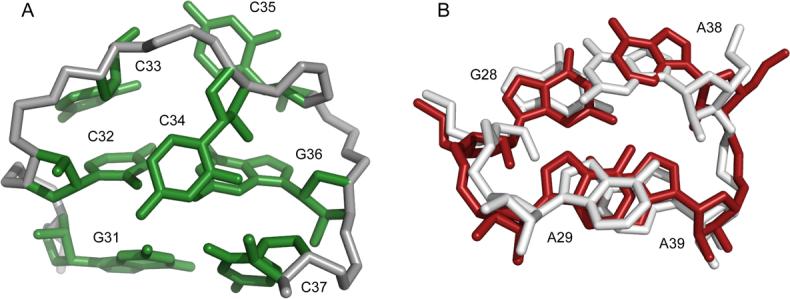
(A) Representative triloop structure with closing G-C pairs. (B) Overlay of the tandem G-A pairs from the 23S rRNA (white) and the G-A sheared pair with cross-stacked adenosine from SIV (red). The bulged residue A30 is not shown for clarity.
The loop contains a modular structural element, the cross-strand adenine stack upon a sheared G-A pair. NOEs that help define the sheared G-A pair are shown (Supplemental Data). A search of RNA structural motifs using the SCOR 2.025 database revealed that G28, A29, A38, and A39 adopt a conformation that is nearly identical to the tandem G-A base-pairs found in D. radiodurans 23S rRNA26 (PDB code 1NKW). The D. rad. structure (white) and the SIV34 structure (red) overlay with an r.m.s.d. of 1.38 Å over all carbon atoms (Figure 7B). While there is no evidence for base-pairing of A29 and A38 in the SIV34 structure, as is seen between A2331 and G2344 in the 23S rRNA, the sequence identity and structural similarities between the two cross-strand stacking motifs are clear.
Why doesn't the SIVSMM RNA structure form a pseudoknot? Pseudoknot formation as depicted in the secondary structure model in Figure 1B is precluded by the length of the single-stranded linker region between the helical end and the loop, which is only 8 nucleotides. The SIVSMM stem-loop structure alone is 52 Å in length, and spans more than a full helical turn. Since the secondary structure in Figure 1B includes an additional lower helix (separated by a C-C mismatch of unknown geometry), it is likely that the 8 nucleotide linker would have to span a distance of > 60 Å, which is physically impossible. The two constructs that we have studied that could potentially form pseudoknots have single stranded linkers that are 10 and 13 nucleotides long (Figure 1C and D, respectively). These linkers are long enough to span the length of the helical turn. Since a translating ribosome paused at the slippery site would be already engaged with the mRNA approximately 13 nucleotides downstream of the first base in the P site (spanning the RNA from U1 to U13 in Figure 1A),27,28 the 4 base-pair helix at the base of the stem would be expected to be at least partially unwound prior to the frameshift event. This condition is mimicked in the SIV14−70 and SIV17−70 constructs (Figures 1C and D), yet these constructs do not form detectable pseudoknot interactions. Thus, we conclude that the loop structure precludes pseudoknot formation by sequestering its nucleotides within a stable (Tm = 66°C) structure. Interestingly, a recently developed RNA secondary structure prediction algorithm capable of predicting pseudoknots29 predicts that the stem-loop structure is more energetically favorable than the pseudoknot fold, and also predicts that the linker is capable of folding into an alternative three base-pair stem-loop structure that is incompatible with pseudoknot formation (data not shown).
SIVSMM frameshift inducing structure is likely similar to HIV-2, but differs from HIV-1
A comparison of SIVSMM, HIV-2 and HIV-1 sequences (www.hiv.lanl.gov) revealed that SIVSMM and HIV-2 have nearly identical sequences for the frameshifting element, but they are both substantially different from the HIV-1 sequence (Figure 8A). Additionally, when the differences between SIVSMM and HIV-2 are mapped to the SIV secondary structure, the only significant difference is observed as a single cytidine-to-uridine transition in the loop (Figure 8B), as all other sequence changes preserve the same base-pairing shown in Figure 1A (data not shown). Modeling and energy minimization of the HIV-2 loop structure from SIV starting structures revealed that the HIV-2 loop can adopt an identical structure, and may be further stabilized by an additional hydrogen-bond between the O4 of U34 and the 2'OH group of C32, as compared to the SIV loop (Figure 8C).
Figure 8.
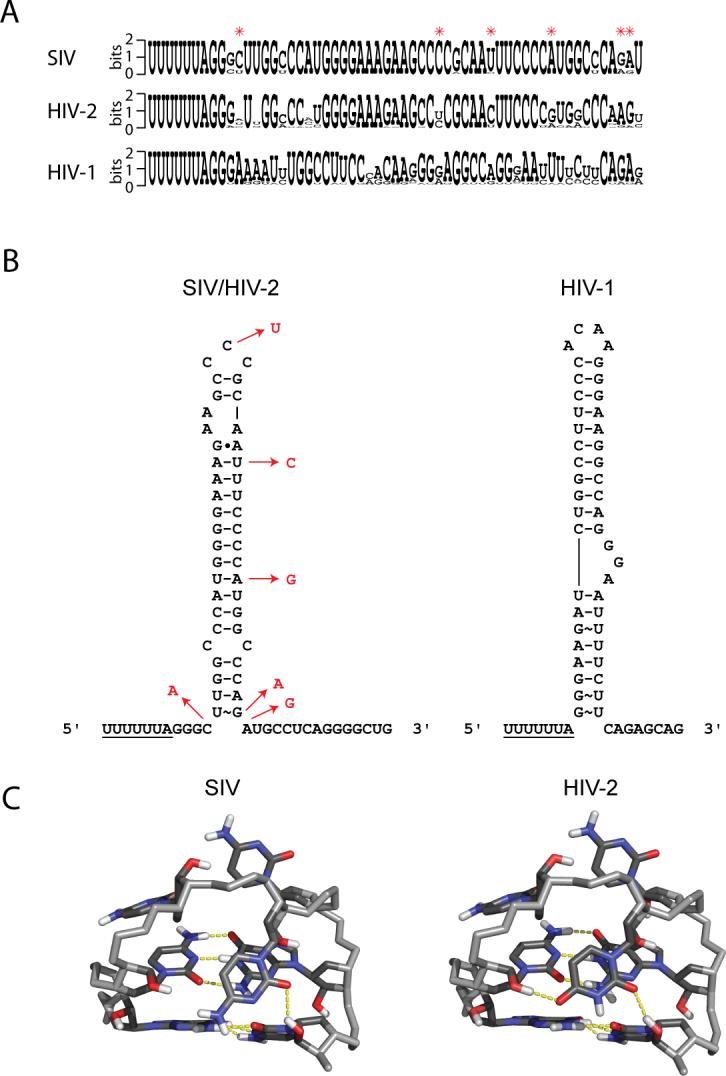
Sequence comparison and analysis of SIV, HIV-2 and HIV-1 frameshift-inducing sequences. (A) Primary sequence comparison of the three viruses. Differences between the SIV and HIV-2 sequences are indicated by red asterisks. Sequence logos were created using WebLogo56 (http://weblogo.berkeley.edu/logo.cgi). (B) Secondary structures of SIV/HIV-2 and HIV-1. Differences between SIV and HIV-2 are indicated by nucleotides in red. HIV-1 structure is shown for comparison. (C) Structure of the CCC (SIV) triloop and model of the CUC (HIV-2) triloop with closing G-C pairs. Potential hydrogen-bonds are shown in yellow.
The frameshift site RNA structure is coupled to the slippery sequence
Most viral RNAs use pseudoknot structures to promote framehshifting. It has been proposed that pseudoknots promote frameshifting through their unique folding topologies, which may be poor substrates for the ribosomal helicase.30-34 In addition, both pseudoknots and stable stem-loops are capable of pausing ribosomes at frameshift slippery sites.35,36 A kinetic pause likely contributes to frameshifting36-38; however, previous studies have found no simple correlation between pausing and frameshifting.35 HIV-1, SIV and HIV-2 are unique in that they utilize highly stable stem-loops for frameshifting.2,7,19,20 We hypothesize that the nature of the frameshift site RNA structure is dependent upon the slippery site to which it is coupled.
All retroviral slippery sites have an XXXYYYZ consensus sequence.39 Previous data indicate that the UUUUUUA slippery sequence used by HIV-1, HIV-2 and SIV is one of the most efficient in promoting programmed −1 translational frameshifting, suggesting that it is more “slippery” than other XXXYYYZ sequences.40 HIV-1, HIV-2 and SIV may only require a thermodynamically stable stem-loop, rather than a pseudoknot, due to the inherent “slipperiness” of the UUUUUUA sequence. For these viruses, the stable stem-loops are sufficient to pause ribosomes and induce the appropriate level of frameshifting. Other viruses, such as the mouse mammary tumor virus (MMTV)32,41 or infectious bronchitis virus (IBV)35,36 may require pseudoknot structures to attain the appropriate level of frameshifting because their slippery sequences (AAAAAAC for MMTV and UUUAAAC for IBV) are not as slippery.42 In these cases, pausing alone may not be sufficient, and pseudoknots may be required because they are stereochemically mismatched substrates for the helicase.43,44
Materials and Methods
RNA synthesis and purification
Milligram quantities of RNA suitable for NMR and UV spectroscopy methods were transcribed in vitro using purified His6-tagged T7 RNA polymerase, synthetic DNA oligonucleotides (Integrated DNA Technologies, Inc.), and NTPs (Sigma-Aldrich). The first two base pairs of the SIV17−50 sequence were flipped relative to wild-type to optimize transcription yields. RNA was purified by denaturing 15% (SIV17−50 RNA) or 8% (SIV17−70 and 14−70 RNA) polyacrylamide gel electrophoresis, identified by UV absorbance, and excised from the gel. RNA was recovered by diffusion into 0.3 M sodium acetate (pH 5.0), precipitated with ethanol, purified on a 6 mL DEAE anion exchange column, again precipitated with ethanol, and desalted on a 15 mL G-25 gel filtration column. The purified RNA was lyophilized, resuspended in water, and brought to pH 7.0 by the addition of 1 M NaOH or by buffer exchange into 10 mM sodium phosphate (pH 7.0). 13C/15N-labelled RNA samples for NMR were prepared using 13C/15N-labelled NTPs (Silantes GmbH, München, Germany). Fully 13C/15N-labelled adenosine/uridine and guanosine/cytidine samples were prepared. A shorter RNA construct of 24 nucleotides (SIV24; corresponding to G22 through C45, Figure 1) was also prepared using NTPs which were specifically deuterated at all positions except the ribose H1'and H2' and base H6/H8 atoms (Cambridge Isotope Laboratories, Inc.). Partial alignment of RNA for RDC measurements was achieved by adding 17 mg/mL of Pf1 filamentous bacteriophage (ASLA Ltd., Riga, Latvia) to the 13C/15N-labelled samples.
In vitro frameshifting assay
Plasmid construction
The sequences investigated here are derived from the most commonly occurring sequence of all SIVSMM RNA isolates analyzed (www.hiv.lanl.gov) and represent the consensus sequence. For each sequence tested, complementary synthetic oligonucleotides (Integrated DNA Technologies, Inc.) with BamH I and Sac I compatible ends were cloned into the p2luc plasmid using the unique BamH I and Sac I sites between the rluc and fluc reporter genes. Oligonucleotidess SIV51 (5'-GATCCTTTTTTAGGCCTTGGTCCATGGGGAAAGAAGCCCCGCAATTTCCCATGGCGAGCT-3'), SIV57 (5'-GATCCTTTTTTAGGCCTTGGTCCATGGGGAAAGAAGCCCCGCAATTTCCCCATGGCTCAAGTGCAGCT-3'), SIV63 (5'-GATCCTTTTTTAGGCCTTGGTCCATGGGGAAAGAAGCCCCGCAATTTCCCCATGGCTCAAGTGCATCAGAGCT-3'), SIV69 (5'-GATCCTTTTTTAGGCCTTGGTCCATGGGGAAAGAAGCCCCGCAATTTCCCCATGGCTCAAGTGCATCAGGGGCTGAGCT-3'), and SIV78 (5'-GATCCTTTTTTAGGCCTTGGTCCATGGGGAAAGAAGCCCCGCAATTTCCCCATGGCTCAAGTGCATCAGGGGCTGATGCCAACGAGCT-3') and their complements were phosphorylated, annealed, and ligated into the p2luc vector to produce the experimental constructs. Positive control sequences and their complements were also cloned into p2luc and have two thymidine residues (bold) in the slippery sequence (underlined) replaced with cytidines and an additional nucleotide inserted immediately before the Sac I complementary sequence (GAGCT). Resultant products were transformed into E. coli competent cells (JM109, Promega). Plasmid DNA was purified from cell cultures (Qiagen) and the sequences of all constructs were verified (University of Wisconsin Biotechnology Center).
In vitro transcription
Purified plasmid DNA was linearized by digestion with Pml I prior to in vitro transcription. RNA (∼3 kb) was transcribed using the T7 RiboMAX Express Large Scale RNA Production System (Promega), the DNA template was removed by treatment with RQ1 RNase-free DNase, and the mixture was extracted with phenol:cholorform 5:1 (pH 4.7) and chloroform:isoamyl alcohol 24:1 (Sigma-Aldrich). The aqueous phase was run on a 5 mL P6 gel filtration column (Bio-Rad) to remove small abortive products and unincorporated NTPs. The purified RNA was lyophilized, resuspended in water, analyzed by agarose (1%) gel electrophoresis, and stored at −80 °C.
In vitro translation
RNA samples were translated in vitro using the Flexi Rabbit Reticulocyte Lysate System (Promega). Translation reactions contained 1 μg of RNA per 10 μL of reaction mixture, 1−2 mM Mg2+, 70 mM KCl, and 20 μM of each amino acid except methionine and leucine (10 μM each). Reactions were incubated at 30°C for 90 minutes.
Frameshift efficiency measurements
Luminescence was measured using a Veritas microplate luminometer equipped with dual-injectors (Turner Biosystems). For each sequence length investigated, 10 replicates of the experimental sequence and 5 replicates of the positive control sequence were examined using the Dual-Luciferase Reporter Assay System (Promega). Luminescence readings were taken for both reporters 10 sec after their respective substrates were injected into the reaction mixture (2 sec lag time prior to measurement). Ratios of firefly/Renilla luminescence were calculated for each of the experimental and control sequences, and replicates were averaged. The ratio of averages (experimental/control) was then determined, which corresponds to the frameshift efficiency (Figure 2). Measurement error was propagated through these calculations to yield a final standard error mean (SEM) value for each of the determined frameshift efficiencies (Figure 2).
UV Spectroscopy
Thermal stability studies were conducted on purified RNA constructs using a Cary Model 100 Bio UV-visible spectrophotometer equipped with a Peltier heating accessory and temperature probe. All samples contained 10 mM sodium phosphate buffer (pH 7.0), 200 mM KCl, and approximately 2 μM RNA. Samples were slowly heated from 20°C to 95°C at a rate of 1°C/min and absorbance data were collected at 260 nm in 1°C increments. Thermal denaturation was determined to be performed under equilibrium conditions, as evidenced by the fact that no hysteresis was observed and similar data were obtained when samples were slowly cooled from 95°C to 20°C. A minimum of three scans were taken for each sample. The data were derivatized, linear baseline corrected, smoothed over a 3°C window, normalized (with the largest value set to 1), and fit using nonlinear least-square fit to solve for Tm and ΔH as previously described.45 Data analysis was performed using Prism 4.0 (GraphPad).
NMR Spectroscopy
All NMR spectra were obtained on Varian Inova (800 and 900 MHz) and Bruker Avance DMX (500, 600 and 750 MHz) spectrometers at the National Magnetic Resonance Facility at Madison (NMRFAM). Spectrometers were equipped with either a conventional proton, carbon, nitrogen (HCN) room temperature probe or a single z-axis gradient HCN cryogenically cooled probe. Exchangeable resonances were assigned by reference to 2D NOESY (150 msec mixing time), 1H-15N HMQC and 2D HNN-COSY spectra of the RNA collected in 90% H2O/10% D2O, pH 7.0 at 283 K. Water suppression for these experiments was achieved by using a 1−1 spin echo or watergate pulse sequence. Non-exchangeable resonances were assigned by reference to 2D NOESY (50, 100, 150, 200, 250, and 300 msec mixing times), 2D 1H-1H TOCSY, 2D 1H-13C HSQC, 3D 1H-13C-1H HCCH TOCSY, and 3D 1H-13C-1H HCCH COSY spectra of the RNA collected in 99.99% D2O, pH 7.0 at 308 K.
Hydrogen-bonds were detected for adenosine/uridine and guanosine/cytidine 13C/15N-labeled samples in 90% H2O/10% D2O, pH 7.0 at 283 K.46 NOESY experiments on the specifically deuterated SIV17−50 RNA were performed to aid in assignment of highly overlapped resonances.
Residual dipolar couplings (RDCs) were measured using J-modulated 1H-13C CT-HSQC experiments (18 planes) in both isotropic and partially oriented 13C/15N-labeled RNA samples in 90% H2O/10% D2O at 308 K. Peak intensities from each plane were fit to yield the 1JC-H coupling values, and RDCs were calculated as the difference between 1JC-H values for isotropic and partially aligned samples.47 PALES software (http://spin.niddk.nih.gov/bax/software/PALES)48 was used to estimate the values for the axial (Da) and rhombic (R) components of the alignment tensor from converged, low energy structures calculated using XPLOR-NIH49 in the absence of RDCs. A subsequent grid search procedure50 performed around the estimated Da and R was used to optimize the values, using lowest overall energies as a target. Optimal Da/R values were determined to be −57/.23 for the stem alignment tensor. RDC values for the loop could not be fit to the same alignment tensor as the stem, and were therefore not used for final structure calculations.
Thermal unfolding of the SIV17−50 RNA (0.8 mM, 90% H2O/10% D2O, pH 7.0) was monitored by collecting a series of 1D 1H spectra at increasing temperature. The temperature was increased from 5 °C to 80 °C in 5 °C increments. All spectra were collected with 32 scans. All data were processed using XwinNMR (Bruker) or NMRPipe51 software, and assignments were made using Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Structure Calculations
NOE distance restraints were categorized as very strong (1.8−2.5 Å), strong (2.0−4.0 Å), medium (2.5−4.5 Å), or weak (3.0−7.0 Å) based on the integrated peak volumes obtained from 2D NOESY spectra (50, 150, 200, 250, and 300 msec mixing times). Torsion angle restraints (angles: α, β, γ, δ, ε, ζ, η, ν1, ν2 and χ) for residues in the stem (G17-A27, U40-C50) were constrained to A-form values52 (±15°), which were consistent with NOESY, TOCSY, HNN-COSY, and RDC data. Additionally, A-form phosphate backbone restraints for these regions were incorporated as previously described.53 Torsion angles for residues in the loop (G28-A39) were left unrestrained, with the exception of experimentally validated glycosidic (χ) and sugar pucker angles (ν1, ν2). Residues with strong H1'-H2' and H1'-H3' cross-peaks (C33, C34, C35) in 2D 1H-1H TOCSY (40 msec mixing time) spectra were restrained to the C2'-endo range (ν1: 25 ± 15°, ν2: −35 ± 15°), residues without H1'-H2' cross-peaks (G28,A29,A30,G31,C37,A39) were restrained to the C3'-endo range (ν1: −25 ± 15°, ν2: 37 ± 15°), and residues which displayed an intermediate H1'-H2' cross-peak (C32, G36, A38, C50), indicative of an averaging of sugar puckers, were left unrestrained. Analysis of the peak volumes of internucleotide and intranucleotide H1'-H6/H8 and H2'-H6/H8 NOEs in 2D NOESY (50 msec mixing time) revealed that C35 was in the syn range; therefore, χ was restrained to 30 ± 40°, as previously observed for a syn-cytidine.24 All other residues in the loop were determined to be in the anti range, and were loosely restrained relative to their sugar pucker conformation (χC3'-endo: −160 ± 30°, χC2'-endo: −125 ± 30°, all other χ angles: −140 ± 35°). Hydrogen-bond distance restraints were used for all base-pairs identified by NOESY and HNN-COSY experiments, and very weak planarity restraints (1 kcal mol−1 Å−2) were enforced during structure calculations.
An extended structure generated in CNS 1.154 was used to calculate 100 starting structures from random initial velocities using distance (NOE), dihedral, hydrogen bonding, and planarity restraints. Structures were subjected to 60 psec (15 fsec time steps) of restrained molecular dynamics in torsion angle space, 90 psec of slow cooling, and 30 psec (5 fsec time steps) of restrained molecular dynamics in Cartesian coordinate space. These structures were then refined with additional RDC restraints using XPLOR-NIH.49 Structures were heated to 3000 K and cooled to 100 K (50 K steps) for 28 psec of restrained molecular dynamics in Cartesian coordinate space, followed by 500 steps of energy minimization using the Powell algorithm and simulated annealing.
Structures were accepted based on criteria of lowest overall energy and lack of NOE violations >0.5 Å and dihedral violations >5°. Structures were viewed and analyzed using MOLMOL55 and figures were created using PyMol (http://www.pymol.org). The final structures were used to back-calculate RDC values using PALES,48 and the r.m.s.d. for observed versus back-calculated RDCs was found to be 1.46 Hz (Table 1).
Modeling the HIV-2 Structure
Modeling of the HIV-2 loop structure was performed using Sybyl 7.3 (Tripos, Inc.). SIV starting structures were modified by replacement of the C34 base with a uracil. Charges were added by the Gasteiger-Huckel method (Tripos), followed by 500 steps of energy minimization using the Powell algorithm and the Tripos force field. Charge addition and energy minimization were also performed on SIV starting structures for comparison. Resulting structures were analyzed using Pymol (http://www.pymol.org), and potential hydrogen-bonds (within 2.0 Å) are displayed (Figure 8).
Data Bank accession codes
Coordinates for the SIV frameshift-inducing RNA have been deposited into the RCSB Protein Data Bank (accession code 2JTP). NMR chemical shift assignments and 2D NOESY time domain data have been deposited into BioMagResBank (accession code 15417).
Acknowledgements
We thank Jared Davis, Sarah Pillard, Nicholas Reiter, and Dipali Sashital for helpful discussions. We also thank Raymond Gesteland (University of Utah) for the generous gift of the p2luc plasmid DNA, Marchel Hill and the Palmenberg lab (University of Wisconsin-Madison) for assistance with luminescence measurements, and Marco Tonelli and the staff at the National Magnetic Resonance Facility at Madison (NMRFAM) for assistance with NMR data collection. All NMR studies were carried out at NMRFAM (http://www.nmrfam.wisc.edu/) with support from U.S. National Institutes of Health (NIH) grants P41RR02301 (Biomedical Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment funding for the facility was provided by the University of Wisconsin, the NIH (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture. This work was supported by NIH grant GM072447 to S.E.B. and by NIH Pre-doctoral Training Grant T32 GM08349 to R.J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- 1.Franchini G, Gurgo C, Guo HG, Gallo RC, Collalti E, Fargnoli KA, Hall LF, Wong-Staal F, Reitz MS. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 2.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–3. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 3.Parkin NT, Chamorro M, Varmus HE. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66:5147–51. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J, Morrow CD. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–7. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YG, Maas S, Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–31. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leger M, Sidani S, Brakier-Gingras L. A reassessment of the response of the bacterial ribosome to the frameshift stimulatory signal of the human immunodeficiency virus type 1. RNA. 2004;10:1225–35. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345:127–36. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 9.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol. 2001;75:1834–41. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung M, Patel P, Davis S, Green SR. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J Virol. 1998;72:4819–24. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 2001;66:233–48. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch VM, Dapolito G, Goeken R, Campbell BJ. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr. Opin. Genet. Dev. 1995;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- 13.Sharp PM, Bailes E, Robertson DL, Gao F, Hahn BH. Origins and Evolution of AIDS Viruses. Biological Bulletin. 1999;196:338–342. doi: 10.2307/1542965. [DOI] [PubMed] [Google Scholar]
- 14.Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, Vandamme AM. Tracing the origin and history of the HIV-2 epidemic. Proceedings of the National Academy of Sciences. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F. Chimpanzee Reservoirs of Pandemic and Nonpandemic HIV-1. American Association for the Advancement of Science. 2006 doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ten Dam EB, Pleij CW, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–36. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le SY, Shapiro BA, Chen JH, Nussinov R, Maizel JV. RNA pseudoknots downstream of the frameshift sites of retroviruses. Genet Anal Tech Appl. 1991;8:191–205. doi: 10.1016/1050-3862(91)90013-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staple DW, Butcher SE. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31:4326–31. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudin C, Mazauric MH, Traikia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J Mol Biol. 2005;349:1024–35. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 20.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J Mol Biol. 2005;349:1011–23. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Grentzmann G, Ingram JA, Kelly PJ, Gestaland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 22.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. Rna. 2003;9:1019–24. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch SR, Tinoco I, Jr, Journals O. The structure of the L3 loop from the hepatitis delta virus ribozyme: a syn cytidine. Nucleic Acids Research. 1998;26:980–987. doi: 10.1093/nar/26.4.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. SCOR: a Structural Classification of RNA database. Nucleic Acids Res. 2002;30:392–4. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High Resolution Structure of the Large Ribosomal Subunit from a Mesophilic Eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 27.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–41. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 28.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 29.Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucl. Acids Res. 2007 doi: 10.1093/nar/gkm258. gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–47. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia A, van Duin J, Pleij CWA. Differential response to frameshift signals in eukaryotic and prokaryotic translational systems. Nucleic Acids Research. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamorro M, Parkin N, Varmus HE. An RNA Pseudoknot and an Optimal Heptameric Shift Site are Required for Highly Efficient Ribosomal Frameshifting on a Retroviral Messenger RNA. Proceedings of the National Academy of Sciences. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. The 9-A solution: how mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–74. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michiels PJ, Versleijen AA, Verlaan PW, Pleij CW, Hilbers CW, Heus HA. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J Mol Biol. 2001;310:1109–23. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–40. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol Cell Biol. 2001;21:8657–70. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bidou L, Stahl G, Grima B, Liu H, Cassan M, Rousset JP. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA Soc. 1997 [PMC free article] [PubMed] [Google Scholar]
- 38.Lopinski JD, Dinman JD, Bruenn JA. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol Cell Biol. 2000;20:1095–103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brierley I. Ribosomal frameshifting viral RNAs. J Gen Virol. 1995;76:1885–92. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 40.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–79. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen LX, Tinoco I,, Jr. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–78. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 42.Plant EP, Dinman JD. Comparative study of the effects of heptameric slippery site composition on- 1 frameshifting among different eukaryotic systems. RNA. 2006;12:666–673. doi: 10.1261/rna.2225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plant EP, Dinman JD. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–33. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–7. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.John DM, Weeks KM. van't Hoff enthalpies without baselines. Protein Science. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grzesiek S, Cordier F, Dingley AJ. Scalar couplings across hydrogen bonds. Methods Enzymol. 2001;338:111–33. doi: 10.1016/s0076-6879(02)38217-x. [DOI] [PubMed] [Google Scholar]
- 47.Ottiger M, Delaglio F, Marquardt JL, Tjandra N, Bax A. Measurement of dipolar couplings for methylene and methyl sites in weakly oriented macromolecules and their use in structure determination. J Magn Reson. 1998;134:365–9. doi: 10.1006/jmre.1998.1546. [DOI] [PubMed] [Google Scholar]
- 48.Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]
- 49.Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 50.Clore GM, Gronenborn AM, Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J Magn Reson. 1998;131:159–62. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 52.Saenger W. Principles of Nucleic Acids. Springer-Verlag; New York: 1984. [Google Scholar]
- 53.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–82. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51, 29–5. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 56.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A Sequence Logo Generator. Cold Spring Harbor Lab. 2004 doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.