Abstract
The shape of enveloped viruses depends critically on an internal protein matrix, yet it remains unclear how the matrix proteins control the geometry of the envelope membrane. We found that matrix proteins purified from Newcastle disease virus adsorb on a phospholipid bilayer and condense into fluidlike domains that cause membrane deformation and budding of spherical vesicles, as seen by fluorescent and electron microscopy. Measurements of the electrical admittance of the membrane resolved the gradual growth and rapid closure of a bud followed by its separation to form a free vesicle. The vesicle size distribution, confined by intrinsic curvature of budding domains, but broadened by their merger, matched the virus size distribution. Thus, matrix proteins implement domain-driven mechanism of budding, which suffices to control the shape of these proteolipid vesicles.
Introduction
Dynamic control of membrane curvature is vital for cells, and protein complexes governing the formation of membrane vesicles use various means of curvature regulation to guide vesicle shape (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006). Apart from coated vesicles formed by highly organized multiprotein complexes (Schekman and Orci, 1996; Matsuoka et al., 1998; Bremser et al., 1999), mechanisms of geometry creation by other, often much simpler protein ensembles are largely unknown. Budding of enveloped viruses is generally governed by only one dedicated matrix protein, though components of intracellular budding machinery have reportedly been involved (Slagsvold et al., 2006). Matrix proteins generally form the tight lining beneath the viral membrane, indicating their direct interactions with the membrane (Garoff et al., 1998). Accordingly, matrix proteins of different viral families have been found to be sufficient to orchestrate membrane budding in cells; their expression and self-assembly on the plasma membrane result in the release of viruslike proteolipid vesicles into the extracellular space (Garoff et al., 1998; Takimoto and Portner, 2004). Thus, matrix proteins directly guide membrane curvature by an internal protein lattice, the topological antipode of conventional protein coats that shape intracellular transport vesicles.
The clustering of membrane-associated proteins that are critically involved in budding (e.g., clathrin) generally results in crystalline ordering (Ford et al., 2001; Kohyama et al., 2003). Correspondingly, polymerization of a solid protein scaffold that enforces a spherical topology on the vesicle membrane remains the most recognized mechanism of vesicle creation to date (Schekman and Orci, 1996; Antonny, 2006). Nevertheless, the mechanisms of curvature creation might be different for vesicles formed by proteins integrated into the vesicle membrane (as opposed to on the membrane, which is common for external protein coats), such as in enveloped viruses or caveolae (Garoff et al., 1998; Sens and Turner, 2004; Bauer and Pelkmans, 2006). In this case, interaction between the lipid bilayer and proteins is generally coupled to membrane curvature (Zimmerberg and Kozlov, 2006), resulting in membrane budding by mere component segregation, as shown in model systems (Simon et al., 1995). Extreme protein crowding on caveolar or viral membranes (Garoff et al., 1998; Sens and Turner, 2004; Bauer and Pelkmans, 2006) also suggests involvement of direct protein–protein interactions in establishing the membrane shape. Yet it remains unclear whether such interactions lead to protein polymerization or the weaker fluid-type protein clustering that has been hypothesized to mediate budding by analogy with fluid lipid domains (Lipowsky, 1992; Dobereiner et al., 1993).
To explore the mechanism of shape creation, we reconstituted membrane budding with purified matrix protein, the key structural component of the envelope of Newcastle disease virus (NDV). As for many paramixoviruses, matrix protein of NDV (M protein) plays a key role in virus formation (Takimoto and Portner, 2004). M protein is absolutely required for viral egression and expression of this protein results in plasma membrane budding and production of viruslike particles by transfected cells (Pantua et al., 2006). The recently reported dependence of NDV formation on lipid rafts (Laliberte et al., 2006), together with experiments showing direct interaction between M protein and pure lipidic membranes (Faaberg and Peeples, 1988; Neitchev and Dumanova, 1992), strongly indicates the synergistic action of M proteins and lipids in the formation of NDV envelopes. We found that the mere interaction of M proteins with the pure lipid bilayer is sufficient to induce self-organization of the proteins into functional budding domains.
Results and discussion
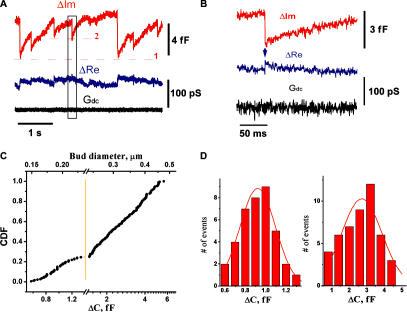
The time-resolved admittance measurements technique, traditionally used to resolve detachment or fusion of small vesicles in cells (Neher and Marty, 1982; Zimmerberg, 1987; Rosenboom and Lindau, 1994; Lollike and Lindau, 1999), was applied to monitor the activity of M protein on the lipid bilayer. We recorded changes in the electrical admittance of a patch, isolated from the planar lipid bilayer (made of a phosphocholine [PC]–phosphoethanolamine [PE]–cholesterol mixture; see Materials and methods) by a small pipette containing 2 μM of M protein. The membrane outside the patch area provided a lipid reservoir to support variations of the patch area. Changes of the imaginary (ΔIm) and real (ΔRe) parts of the admittance were detected ∼1 min after establishing a tight contact between the pipette and the membrane in 7 out of 15 patches (Fig. 1 A). The ΔIm tracing, which tracks changes in the patch area (Lollike and Lindau, 1999), showed periodic variations, with each period consisting of a slow increase followed by a fast decrease of apparent membrane area. Such activity indicates formation of membrane buds (see Fig. 2); during the initial rising stage the membrane area is retrieved from the lipid reservoir into the bud, whereas the fast area drop indicates its detachment. Excision of the membrane patch from the reservoir membrane led to the impairment of the ΔIm alterations and destabilization of the membrane patch, confirming that variations of ΔIm report changes in the patch area, requiring substantial lipid addition (an isolated patch membrane cannot store enough excess area for multiple bud formation). The periodic increases seen in the ΔIm tracing were not accompanied by any substantial changes of the permeability of any part of the membrane within the patch pipette (measured as membrane conductance at constant holding potential [Gdc]; Fig. 1, A and B; Neher and Marty, 1982).
Figure 1.
Interaction of M protein with lipid membrane, monitored by patch clamp admittance measurements. (A) Changes of the admittance (ΔIm and ΔRe) and ionic permeability (Gdc) of the patch connected to the membrane reservoir upon application of 2 μM of M protein. Level 1 shows the background level of ΔIm corresponding to the initial area of the patch. ΔIm deviations back and forth to level 1 indicate reversible changes of the patch area, and each single alteration (e.g., around level 2) indicates a budding event. (B) Expanded selection from black box in A. Transient increase in ΔRe (arrow) indicates formation of a thin membrane neck. (C) Cumulative distribution of the values of ΔIm jumps and the corresponding diameters of the spherical membrane particle. The initial part of the distribution (up to ∼1.3 fF) is expanded to show a Gaussian-like profile. (D) Left histogram shows the distribution of small ΔIm jumps from C; right histogram shows the distribution of ΔIm jumps obtained at elevated (5 μM) concentration of M protein.
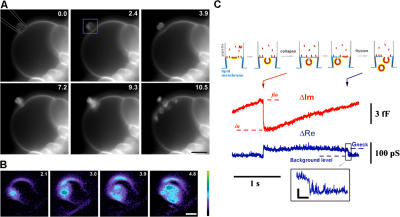
Figure 2.
Visualization of the budding activity of M protein on a membrane patch. (A) Frame sequence (time in seconds) illustrating budding from a patch pipette (approximately drawn in the first image) containing 2 μM of M protein observed on a GUV. A small part of the large GUV, attached to a platinum electrode used for electroformation, was sucked into the pipette. Recording began after establishing a stable contact between the GUV and the pipette. Bar, 5 μm. (B) Expanded images, corresponding to the area marked by the purple rectangle in A, illustrate brightening of the membrane patch upon M protein adsorption. (C) The scheme outlines a correspondence between the changes in ΔIm and the budding. Levels in and fin show the ΔIm increase caused by formation of a bud. Red arrow indicates bud closure; blue arrow indicates fission of the neck. (inset) The fission shown in detail. Bars: (A) 5 μm; (B) 1 μm; (C, horizontal) 40 ms; (C, vertical) 20 pS.
Sharp drops of ΔIm (Fig. 1 A, B) were often followed by transient rises of ΔRe, illustrating formation of a thin neck connecting the bud and membrane patch, as during the pinching-off of an endosome in a cellular system (Rosenboom and Lindau, 1994, Suss-Toby et al., 1996; Frolov et al., 2003). The amplitude of the ΔRe increase was usually much smaller than the one of the preceding ΔIm drop (Fig. 1 B), thus the value of the ΔIm jump approached total electrical capacitance of the bud membrane (see Materials and methods) proportional to the bud area. The cumulative distribution function of the values of ΔIm jumps is rather broad and skewed, with a pronounced singularity at ∼1.3 fF (188 jumps in total; Fig. 1 C). This singularity breaks the distribution into two parts. Smaller jumps (Fig. 1 C, left of the yellow line) have normal size distribution, with a mean value of 0.92 ± 0.17 fF (SD, n = 40; Fig. 1 D, left), corresponding to a membrane area of ∼0.1 μm2 (with specific capacitance of 10 fF/μm2). The diameter of a spherical bud of such area is ∼180 nm, close to the typical sizes of an NDV particle (150–300 nm; Takimoto and Portner, 2004). Distribution of the larger jumps is close to log-normal, ranging from 200 to 500 nm consistently with the size heterogeneity of viruslike particules produced by M protein (Pantua et al., 2006). Increasing the M protein concentration in the pipette to 5 μM led to an overall increase of the values of ΔIm jumps to 2.7 ± 1.1 fF (SD, n = 47; Fig. 1 D, right).
To directly assay shape transformations of the membrane patch, we visualized the activity of M protein on the membrane of giant unilamellar vesicles (GUVs; PC–PE–cholesterol mixture) containing a fluorescent lipid probe. A small patch of GUV membrane was isolated inside a pipette containing M protein. As in admittance measurement experiments, the membrane outside the patch area provided a lipid reservoir to support budding. Fig. 2 A demonstrates that shortly after establishing a stable contact between a GUV membrane and a pipette containing 2 μM of M protein, the fluorescence of the membrane patch inside the pipette increased sharply as the proteins adsorbed on the membrane (Fig. 2 B). The subsequent membrane rearrangements resulted in formation of round vesicles of different diameters visible near the patch, confirming the assumption on the spherical topology of the buds. The vesicles' sizes are more broadly distributed and generally larger than those observed on the planar lipid bilayer, likely because of differences in lateral tension for each lipid system (Sens and Turner, 2004). Unidirectional budding of multiple vesicles demonstrates that the adsorbed proteins impose negative curvature on the membrane (here defined as the mean curvature of the membrane monolayer covered by proteins). With retrieval of the membrane area into the vesicles, the GUV diameter was progressively decreasing; thus, contact with the pipette did not interfere with lipid exchange between the external reservoir and the patch membrane. Finally, the GUV membrane detached from the pipette and multiple vesicles were seen moving inside the GUV (Fig. 2 and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1).
The moments of vesicle detachment were also resolved by admittance measurements. The scheme in Fig. 2 C illustrates the complete sequence of membrane budding and fission. First, the membrane bud closed (Fig. 2 C, red arrow), reflecting the abrupt narrowing of the membrane neck connecting the bud and membrane patch (Lipowsky, 1992; Frolov et al., 2003). Afterward, the neck conductance (proportional to ΔRe; see Materials and methods) dropped below the level of resolution, indicating membrane fission (Fig. 2 C, blue arrow). This final drop was detected in a small fraction of trials (∼2%). Generally, ΔRe steadily decreased below the level of resolution (Fig. 1 B), likely because of the gradual elongation and/or thinning of the neck. Nevertheless, appearance of freely moving intralumenal vesicles (Fig. 2 A) corroborates the ultimate fission of the vesicle necks.
Intralumenal vesicles were also efficiently formed when 4 μM of M protein was applied from a thin pipette and placed near a GUV by a weak pulse of positive hydrostatic pressure. Shortly after the protein application, changes in membrane fluorescence as well as membrane deformations were detected. They initially appeared as bright domains and invaginations associated with the GUV membrane (Fig. 3 A and see Fig. 5), and then transformed into intralumenal vesicles moving inside the original GUV (Fig. 3, A and B; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1).
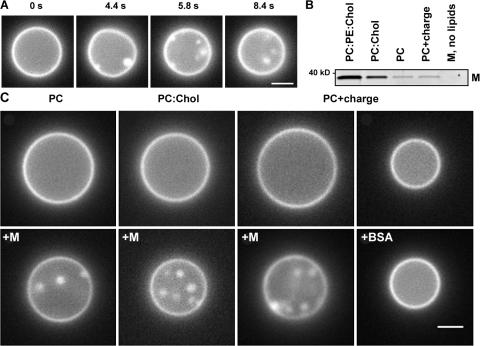
Figure 3.
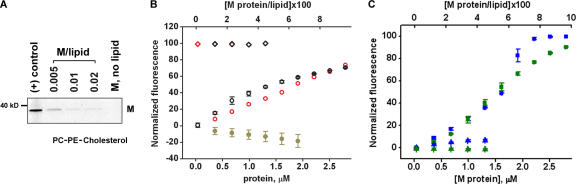
Formation of intralumenal vesicles by M protein applied to GUVs of different lipid compositions. (A) Frame sequence shows formation of intralumenal vesicles after M protein application (at 0 s) to GUV (PC–cholesterol mixture). (B) M protein adsorption on LUVs of different lipid compositions (0.005 protein/lipid ratio) measured by gradient flotation technique. The same protein concentration for all bands was loaded and the control fraction (M, no lipids) was taken at the same level as the liposome fraction. (C) Effect of M protein and BSA application (4 μM in the delivery pipette) on the morphology of GUV of different lipid compositions. Images were taken before (top) and ∼2 min after (bottom) protein application. Representative images of three independent experiments are shown. Bars, 2 μm.
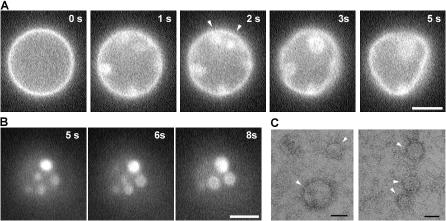
Figure 5.
Formation of membrane domains after M protein application to GUVs. (A) Changes of membrane fluorescence and deformations of GUVs (PC–PE–cholesterol) induced by M protein (added at t = 0). Arrowheads show joining of bright domains. (B) Bright spots merger on GUV flattened on the glass surface. (C) Negative staining of M proteins condensing on a lipid monolayer (arrowheads). Bars: (A and B) 5 μm; (C) 50 nm.
Vesicle formation was stimulated by cholesterol and membrane charge (Fig. 3 C). We compared the efficiency of M protein binding to large unilamellar vesicles (LUVs) containing different amounts of cholesterol and charge lipids. For all lipid compositions tested, a fraction of M proteins bound tightly to the LUV membrane (Fig. 3 B). The binding efficiency was not affected by the membrane charge (Fig. 3 B), corroborating earlier findings that M protein adsorption on the lipid bilayer is predominantly nonelectrostatic (Faaberg and Peeples, 1988). Thus, charge lipids enhance the budding activity of already bound M proteins. Cholesterol, however, stimulates both adsorption and budding activity of M protein. Notably, with addition of 30 mole fraction × 100 (mol%) of cholesterol, which doubles the bending rigidity of the GUV membrane (Henriksen et al., 2004), the budding efficiency of M protein was not diminished but rather augmented (Fig. 3 C). This finding demonstrates that cholesterol, an abundant component in the NDV membrane, can actively participate in the virus budding (Laliberte et al., 2006). Interestingly, the presence of PE also augments protein adsorption (Fig. 3 B) and supports effective membrane budding (Fig. 2 A), likely through its intrinsic negative curvature.
Overall, vesicle formation and changes of membrane fluorescence were detected with four different batches of M protein (18 experiments total). No comparable changes were observed on GUVs perfused with the buffer containing no protein or 4 μM BSA (Fig. 3 C, right; and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1). We conclude that through interactions with the lipid bilayer, M protein implements the genetically encoded information required to create virus geometry.
To gain insight into the mechanism of curvature creation, we analyzed structural alterations in the lipid bilayer induced by M protein. Such alterations, correlated with membrane deformations, were first evident from the increase of fluorescence of membrane patches during vesicle budding (Fig. 2 B). A similar increase of membrane fluorescence is induced when M protein binds to LUV (Fig. 4, A and B), whereas BSA caused no effect at comparable concentrations (Fig. 4 B). Adsorption of M protein to LUV induced comparable dequenching of two different fluorescent probes, rhodamine (Rh)–dioleoyl-PE (DOPE) and boron dipyrromethane difluoride (BODIPY)–Gm1, but did not alter the fluorescence of LUV containing nonquenched dyes (Fig. 4 B). The similar behavior of two chemically different fluorophores and the lack of influence of proteins on nonquenched dyes preclude specific interactions between the fluorophores and the protein. Furthermore, the increase of steady-state anisotropy of the BODIPY-Gm1 fluorescence upon M protein addition was detected for both quenched and nonquenched dye (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1), suggesting that membrane-associated proteins impose general constraints on lipid mobility (Neitchev and Dumanova, 1992).
Figure 4.
Interaction of M protein with LUVs. (A) M protein adsorption on PC–PE–cholesterol LUVs at different protein/lipid ratios measured by gradient flotation. The same protein concentration was loaded for all bands. The control fraction (M, no lipids) was taken at the same level as the liposome fraction. Positive control shows the M protein band. (B) Sequential additions of 0.3 μM of M protein or BSA to LUV caused dequenching of Rh-DOPE or BODIPY-Gm1 fluorescence (red and black circles). No changes were detected for nonquenched dyes (red and black diamonds) or when BSA was added (dark yellow circles). (E) The same additions of M protein induce the release of LUV-entrapped ANTS/DPX (blue squares) or 70-kD FITC-conjugated dextrans (green squares), seen as changes of normalized fluorescence intensity. The addition of the same amount of the protein mixed with α-chymotrypsin 1:5 causes minor release of ANTS/DPX and dextrans (blue and green triangles). Bars show SD.
Changes in membrane fluorescence of LUV were detected only at relatively high protein concentrations sufficient to produce membrane deformation (see Fig. 1). At those concentrations, we detected leakage of contents from LUV loaded with aqueous fluorescent markers, either small (8-aminonaphthalene–1,3,6–trisulfonic acid/p-xylene-bis-pyridium bromide [ANTS/DPX]) or large (70 kD FITC-dextran). Proteolytic treatment of M protein greatly impaired the release efficiency (Figs. 4 C and S2, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1). Release efficiency was comparable for both markers for the same amount of the protein added (Fig. 4 C), in agreement with vesicle bursting. Previously, we established that M proteins did not form any conductive pathways in the lipid bilayer, such as proteolipid pores (Fig. 1, Gdc tracings). Rather, the vesicles' rupture indicated membrane deformations induced by the protein. As liposome volume can be considered fixed at short time scales, substantial membrane deformations (e.g., membrane invaginations) tend to increase the surface/volume ratio of a liposome, thus stretching and ultimately rupturing the liposome membrane as in experiments on the osmotic rupturing of LUVs (Mui et al., 1993). Thus, the bending of the lipid bilayer by M protein is generally correlated with an increase in both intensity and anisotropy of membrane fluorescence.
The relatively high protein concentration required to reconstitute M protein activity suggests that protein condensation in budding areas is the likely cause of fluorescence intensity. Indeed, viral M proteins assemble into a tight layer under the viral envelope and also can aggregate in vitro (Faaberg and Peeples, 1988). Here, the experiments on GUV containing Rh-DOPE in a self-quenched concentration directly demonstrate formation of distinct membrane domains. Shortly after protein application to GUV, bright spots formed within the original GUV contour (Fig. 5 A). The spots enlarged and merged as the GUV quickly deformed away from its initially spherical shape (Fig. 5 A and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1). Some bright spots appeared as budlike membrane invaginations, similar to those observed with the M protein of vesicular stomatitis virus (Solon et al., 2005). On deflated GUVs flattened on the coverslip, the bright spot either budded away as small vesicles or continued growing and merging in large circles (Fig. 5 B and Video 5), which is behavior that has been previously described for fluidlike lipid domains (Samsonov et al., 2001; Laradji and Sunil Kumar, 2005; Yanagisawa et al., 2007). The likely cause for the fluorescence dequenching in the domain areas is limitation of lipid mobility by membrane-associating M proteins (Neitchev and Dumanova, 1992), which would impede energy exchange between the fluorophores.
Self-assembly of M proteins into circular domains on the lipid surface was further confirmed by EM observations. Circular patterns were detected after M protein adsorption on a lipid monolayer preformed on the air–water interface (Fig. 5 C; Ford et al., 2001). No such objects were detected in control experiments when only M protein or lipids were applied (not depicted). Though a circular shape (Fig. 5 C) is a characteristic of fluids, similar patterns have also been detected for polymerized protein coats whose shape is defined by the polymerization pattern (Ford et al., 2001). However, growing via merger that gives rise to a wide difference in domain sizes (from submicrometer clusters to micrometer-sized domains; Fig. 5 B), in striking difference to the well-defined size of protein lattices (Ford et al., 2001), requires internal fluidity of the domains. Thus, the generic tendency of M protein to self-aggregate (Faaberg and Peeples, 1988; Sagrera et al., 1998) is moderated on the membrane so that fluidlike proteolipid domains form.
The dynamics of vesicle formation observed by admittance measurements are consistent with the domain-driven mechanism of budding, originally proposed for fluid lipid domains (Lipowsky, 1992). Growing lipid domains destabilize and collapse into a closed vesicle, which might still remain attached via a thin neck, when the energies of both become comparable, closely resembling vesicle formation by M proteins (Fig. 1 B, arrow). The subsequent vesicle separation is triggered through instabilities in the domain boundary (Lipowsky, 1992; Dobereiner et al., 1993) and doesn't require the participation of specialized fission proteins. A domain merger could account for large deviations in the size of vesicles produced by M protein; although the smaller vesicles would represent domains budding independently (Fig. 1 D, left), the larger vesicles result from a domain merger.
The formation of vesicles from fluid domains in a planar bilayer with high lateral tension σ (typically σ is ∼10−3 N/m2; Frolov et al., 2003) requires substantial energy to pull lipid material from the reservoir and bend it into a sphere. For a 100-nm vesicle, such energy would reach several thousand kBT, where kB is Boltzmann's constant and T is the temperature in degrees Kelvin (ΔF is ∼8πkc+ σS, where S is the vesicle area and the bending modulus kc is ∼20 kBT). However, if M proteins are as tightly packed on the vesicle membrane as inside the virus, the number of proteins per 100-nm vesicle is >1,000 (e.g., at 0.05 protein/lipid ratio on the membrane surface; Fig. 3, B and C). At such densities, the energy cost to pull material and bend it into a sphere per protein is low (approaching 1 kBT). Thus weak interactions between proteins and lipids in the domains can combine to provide enough energy for curvature creation. This estimation corroborates the notion that the weak association of M proteins on the membrane can energetically support membrane deformations.
Besides providing the required energy, the same association of M proteins controls membrane geometry, producing membrane vesicles of the desired shape. Long-range coordination of membrane deformations required for vesicle formation is based not on the intrinsic topology of the protein lattice but on proteolipid interactions within the fluidlike budding domain. These interactions are manifested as intrinsic curvature of the domain, which is evident for unidirectional vesicle budding (Figs. 2 A and 3 C) and the line tension of the domain boundary. Both factors drive membrane curvature, creating viruslike membrane vesicles from the pure lipid bilayer. Although fluid domain–driven budding is generally sensitive to various membrane parameters (Lipowsky, 1992, Dobereiner et al., 1993, Laradji and Sunil Kumar, 2005), we demonstrated that vesicle populations with a narrow size distribution indeed can be obtained (Fig. 1 D; Dobereiner et al., 1993; Sens and Turner, 2004). Thus, despite its intrinsic simplicity, weak protein condensation on a membrane surface provides a powerful tool to regulate membrane shape and topology.
Materials and methods
M protein, lipid compositions, and ionic buffers
M protein was purified from the “clone 30” strain of NDV as described previously (Garcia-Sastre et al., 1989), with 5 mM Ca2+ added to all buffers used during purification. The obtained M protein pellet was dissolved in 1 M KCl, 20 mM Hepes, and 0.2 EDTA, pH 7.4. Concentration of the protein was measured by BCA Protein Assay kit (Thermo Fisher Scientific). All of the experiments were conducted in 100 mM KCl, 20 mM Hepes, and 0.2 mM EDTA, pH 7.4 (buffer A). Gm1 ganglioside conjugated with BODIPY-FL (Invitrogen) in the polar head region (BODIPY-GM1) was synthesized as described previously (Samsonov et al., 2001). Dioleoyl-PE (DOPC), 1-palmitoyl-2-oleoyl-PE (POPC), DOPE, 1,2-dioleoyl-phosphoglycerol (DOPG), and DOPE–lissamine Rh B sulfonyl (Rh-DOPE) were obtained from Avanti Polar Lipids, Inc. The following lipid compositions were used (mole ratio is indicated): DOPC/DOPE/cholesterol, 58:28:10 (PC–PE–cholesterol); POPC (PC); POPC/cholesterol, 66:30 (PC–cholesterol); POPC/DOPG, 81:15 (PC + charge). All were supplemented with 4 mol% of Rh-DOPE or BODIPY-GM1. For experiments with nonquenched fluorophores in PC–PE–cholesterol, the amount of Rh-DOPE or BODIPY-Gm1 was decreased to 0.2 mol% and the amount of PC and PE was increased proportionally.
Preparation of liposomes
100-nm LUVs were prepared by extrusion in buffer A or buffer (osmotically balanced with A) containing ANTS/DPX or 70 kD FITC-dextran in self-quenched concentration, as described previously (Basanez et al., 2001). GUVs were prepared by electroformation using platinum wire electrodes (Goodfellow Metals; Angelova and Dimitrov, 1988). The electroformation was performed in sucrose buffer, osmotically equilibrated with buffer A. The resulting GUVs were either detached from the electrode and put in buffer A or left on the electrode and perfused with buffer A.
Protein binding to LUV
5 μM of M protein was incubated for 5 min with LUVs of different lipid compositions at different protein/lipid ratios. The amount of LUV was normalized for the total fluorescence of Rh-DOPE incorporated. The LUV fraction was separated from unbound protein using the Ficoll gradient flotation method (Fraley et al., 1980) and analyzed by SDS-PAGE using SYPRO Ruby protein gel stain (Invitrogen).
Fluorescence measurements
Leakage of ANTS or FITC-dextran and changes of fluorescence intensity of Rh-DOPE or BODIPY-Gm1 after addition of the M protein to LUV was determined at ambient temperature by spectrofluorimetric measurements using a luminescence spectrometer (Aminco-Bowman SLM-2; Spectronic Instruments, Inc.). The normalized fluorescence intensity Fn was recalculated from integral fluorescence intensity of LUV as follows: Fn = (F−Fi)/(Ff−Fi), where Fi corresponds to F before the protein addition and Ff – to F after complete disruption of LUV (infinite dilution of the fluorophores) by detergent (0.1% of Triton X-100; Sigma-Aldrich). 380/520-nm excitation/emission wavelengths were used for ANTS/DPX signal detection, 550/590 nm for Rh-DOPE, 505/525 nm for BODIPY-Gm1, and 490/520 nm for FITC-dextran.
Fluorescent microscopy of M protein–GUV interaction
The visualization of GUVs attached to the electrode was performed on an inverted microscope (Axiovert 200; Carl Zeiss, Inc.) using a 40×, 0.75 NA objective (ACHROPLAN; Carl Zeiss, Inc.). GUVs detached from the electrode were settled on the bottom of a 170-μm-thin glass 35-mm dish. The dishes were preincubated with 1 g/liter BSA for 1 min and thoroughly washed with buffer A to reduce GUV binding to the glass. The interaction of M protein with GUVs detached from the electrode was recorded using Axiovert 200 or Olympus IX-70 inverted microscopes both equipped with 150×, 1.45 NA objectives (Olympus). The images were digitized by CoolSNAP EZ (Photometrics) or an intensified charge-coupled device camera (VE1000SIT; Dage-MTI) connected to IPLab (BioVision) or Metamorph Flashbus (MDS Analytical Technologies), respectively.
Analysis of M protein condensation on lipid monolayer
The analysis technique was adapted from Ford et al. (2001). In brief, PC–cholesterol lipid solution in methanol/chloroform (9:1) was deposited on a buffer droplet. After 1-h equilibration, a carbon-coated gold EM grid (Electron Microscopy Sciences) was placed on top of the buffer droplet where the lipid monolayer has been formed. M protein was applied to the buffer and, after 1-h incubation, the grid was removed and stained with uranyl acetate (2% solution) for further observations with a transmission EM (Tecnai G2; FEI Company).
Admittance measurements
Planar lipid bilayers were prepared by the Mueller-Rudin technique from the PC–PE–cholesterol mixture in squalane and patch clamped as decribed previously (Frolov et al., 2003). Admittance measurements were performed using a patch clamp amplifier (Extracellular Patch Clamp 8; HEKA) and a PC-44 acquisition board (Signalogic) with on-board software lock-in (Ratinov et al., 1998) using a 5,000-Hz, 100-mV sinewave superimposed with −20 mV of holding potential. The bud capacitance ΔC and the neck conductance Gneck were estimated offline (Rosenboom and Lindau, 1994; Lollike and Lindau, 1999): ΔC = (ΔRe2 + ΔIm2)/ΔIm/ω (ω = 2πf; f is the sinewave frequency), if ΔRe ≪ ΔIm jump, thus ΔC ≈ ΔImjump/ω; accordingly, G neck = (ΔRe2 + (ωΔC − ΔIm jump)2)/ΔRe ≈ ΔRe.
Online supplemental material
Online supplemental material describes measurements of the M protein purity and details of the protein enzymatic treatment (Fig. S1), as well as measurements of the steady-state anisotropy of BOPIPY-GM1 fluorescence (Fig. S2). Video 1 shows M protein–driven vesicle formation from a membrane patch isolated from GUVs by a patch pipette. Video 2 shows formation of such vesicles by transient protein application to a GUV, whereas Video 3 shows no effect of BSA application. Videos 4 and 5 show temporal and spatial changes in membrane fluorescence induced by M proteins on GUV. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705062/DC1.
Acknowledgments
Supported by the intramural research program of the National Institute of Child Health and Human Development; Spanish Fondo de Investigaciones Sanitarias grant FIS-PI051796, cofinanced by Fonds Européen de Développement Régional-Fonds Social Européen; and the Spanish Ministerio de Educación y Ciencia Formacíon de Profesorado Universitario program (predoctoral fellowship AP-2004-6065 to J. Ayllon).
I.I. Mikhalyov's present address is Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia.
Abbreviations used in this paper: ANTS/DPX, 8-aminonaphthalene–1,3,6– trisulfonic acid/p-xylene-bis-pyridium bromide; BODIPY, boron dipyrromethane difluoride; DOPE, dioleoyl-PE; GUV, giant unilamellar vesicle; LUV, large unilamellar vesicle; M, matrix protein of NDV; NDV, Newcastle disease virus; PC, phosphocholine; PE, phosphoethanolamine; Rh, rhodamine.
References
- Angelova, M.I., and D.S. Dimitrov. 1988. A mechanism of liposome electroformation. Prog. Colloid. Polym. Sci. 76:59–67. [Google Scholar]
- Antonny, B. 2006. Membrane deformation by protein coats. Curr. Opin. Cell Biol. 18:386–394. [DOI] [PubMed] [Google Scholar]
- Basanez, G., J. Zhang, B.N. Chau, G.I. Maksaev, V.A. Frolov, T.A. Brandt, J. Burch, J.M. Hardwick, and J. Zimmerberg. 2001. Pro-apoptotic cleavage products of Bcl-xL form cytochrome c-conducting pores in pure lipid membranes. J. Biol. Chem. 276:31083–31091. [DOI] [PubMed] [Google Scholar]
- Bauer, M., and L. Pelkmans. 2006. A new paradigm for membrane-organizing and shaping scaffolds. FEBS Lett. 580:5559–5564. [DOI] [PubMed] [Google Scholar]
- Bremser, M., W. Nickel, M. Schweikert, M. Ravazzola, M. Amherdt, C.A. Hughes, T.H. Sollner, J.E. Rothman, and F.T. Wieland. 1999. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 96:495–506. [DOI] [PubMed] [Google Scholar]
- Dobereiner, H.G., J. Kas, D. Noppl, I. Sprenger, and E. Sackmann. 1993. Budding and fission of vesicles. Biophys. J. 65:1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faaberg, K.S., and M.E. Peeples. 1988. Association of soluble matrix protein of Newcastle disease virus with liposomes is independent of ionic conditions. Virology. 166:123–132. [DOI] [PubMed] [Google Scholar]
- Ford, M.G., B.M. Pearse, M.K. Higgins, Y. Vallis, D.J. Owen, A. Gibson, C.R. Hopkins, P.R. Evans, and H.T. McMahon. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 291(5506):1051–1055. [DOI] [PubMed] [Google Scholar]
- Fraley, R., S. Subramani, P. Berg, and D. Papahadjopoulos. 1980. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 255:10431–10435. [PubMed] [Google Scholar]
- Frolov, V.A., V.A. Lizunov, A.Y. Dunina-Barkovskaya, A.V. Samsonov, and J. Zimmerberg. 2003. Shape bistability of a membrane neck: a toggle switch to control vesicle content release. Proc. Natl. Acad. Sci. USA. 100:8698–8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre, A., J.A. Cabezas, and E. Villar. 1989. Proteins of Newcastle disease virus envelope: interaction between the outer hemagglutinin-neuraminidase glycoprotein and the inner non-glycosylated matrix protein. Biochim. Biophys. Acta. 999:171–175. [DOI] [PubMed] [Google Scholar]
- Garoff, H., R. Hewson, and D.J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen, J., A.C. Rowat, and J.H. Ipsen. 2004. Vesicle fluctuation analysis of the effects of sterols on membrane bending rigidity. Eur. Biophys. J. 33:732–741. [DOI] [PubMed] [Google Scholar]
- Kohyama, T., D.M. Krol, and G. Gompper. 2003. Budding of crystalline domains in fluid membranes. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 68:061905. [DOI] [PubMed] [Google Scholar]
- Laliberte, J.P., L.W. McGinnes, M.E. Peeples, and T.G. Morrison. 2006. Integrity of membrane lipid rafts is necessary for the ordered assembly and release of infectious Newcastle disease virus particles. J. Virol. 80:10652–10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laradji, M., and P.B. Sunil Kumar. 2005. Domain growth, budding, and fission in phase-separating self-assembled fluid bilayers. J. Chem. Phys. 123:224902. [DOI] [PubMed] [Google Scholar]
- Lipowsky, R. 1992. Budding of membranes induced by intramembrane domains. Journal de Physique II. 2:1825–1840. [Google Scholar]
- Lollike, K., and M. Lindau. 1999. Membrane capacitance techniques to monitor granule exocytosis in neutrophils. J. Immunol. Methods. 232:111–120. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., L. Orci, M. Amherdt, S.Y. Bednarek, S. Hamamoto, R. Schekman, and T. Yeung. 1998. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 93:263–275. [DOI] [PubMed] [Google Scholar]
- McMahon, H.T., and J.L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438(7068):590–596. [DOI] [PubMed] [Google Scholar]
- Mui, B.L., P.R. Cullis, E.A. Evans, and T.D. Madden. 1993. Osmotic properties of large unilamellar vesicles prepared by extrusion. Biophys. J. 64:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher, E., and A. Marty. 1982. Discrete changes of cell membrane capacitance observed under conditions of enhanced secretion in bovine adrenal chromaffin cells. Proc. Natl. Acad. Sci. USA. 79:6712–6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitchev, V.Z., and L.P. Dumanova. 1992. Effects of the components of Newcastle disease virus on the structural order of lipid assemblies. Mol. Biol. Rep. 16:27–31. [DOI] [PubMed] [Google Scholar]
- Pantua, H.D., L.W. McGinnes, M.E. Peeples, and T.G. Morrison. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J. Virol. 80:11062–11073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratinov, V., I. Plonsky, and J. Zimmerberg. 1998. Fusion pore conductance: experimental approaches and theoretical algorithms. Biophys. J. 74:2374–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenboom, H., and M. Lindau. 1994. Exo-endocytosis and closing of the fission pore during endocytosis in single pituitary nerve terminals internally perfused with high calcium concentrations. Proc. Natl. Acad. Sci. USA. 91:5267–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagrera, A., C. Cobaleda, S. Breger, M.J. Marcos, V. Shnyrov, and E. Villar. 1998. Study of the influence of salt concentration on Newcastle disease virus matrix protein aggregation. Biochem. Mol. Biol. Int. 46:429–435. [DOI] [PubMed] [Google Scholar]
- Samsonov, A.V., I. Mihalyov, and F.S. Cohen. 2001. Characterization of cholesterol-sphingomyelin domains and their dynamics in bilayer membranes. Biophys. J. 81:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman, R., and L. Orci. 1996. Coat proteins and vesicle budding. Science. 271:1526–1533. [DOI] [PubMed] [Google Scholar]
- Sens, P., and M.S. Turner. 2004. Theoretical model for the formation of caveolae and similar membrane invaginations. Biophys. J. 86:2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J., M. Kuhner, H. Ringsdorf, and E. Sackmann. 1995. Polymer-induced shape changes and capping in giant liposomes. Chem. Phys. Lipids. 76:241–258. [Google Scholar]
- Slagsvold, T., K. Pattni, L. Malerod, and H. Stenmark. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 16:317–326. [DOI] [PubMed] [Google Scholar]
- Solon, J., O. Gareil, P. Bassereau, and Y. Gaudin. 2005. Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J. Gen. Virol. 86:3357–3363. [DOI] [PubMed] [Google Scholar]
- Suss-Toby, E., J. Zimmerberg, and G.E. Ward. 1996. Toxoplasma invasion: the parasitophorous vacuole is formed from host cell plasma membrane and pinches off via a fission pore. Proc. Natl. Acad. Sci. USA. 93:8413–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133–145. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, M., M. Imai, T. Masui, S. Komura, and T. Ohta. 2007. Growth dynamics of domains in ternary fluid vesicles. Biophys. J. 92:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg, J. 1987. Molecular mechanisms of membrane fusion: steps during phospholipid and exocytotic membrane fusion. Biosci. Rep. 7:251–268. [DOI] [PubMed] [Google Scholar]
- Zimmerberg, J., and M.M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7:9–19. [DOI] [PubMed] [Google Scholar]