Abstract
The spindle checkpoint delays anaphase onset until every chromosome kinetochore has been efficiently captured by the mitotic spindle microtubules. In this study, we report that the human pre–messenger RNA processing 4 (PRP4) protein kinase associates with kinetochores during mitosis. PRP4 depletion by RNA interference induces mitotic acceleration. Moreover, we frequently observe lagging chromatids during anaphase leading to aneuploidy. PRP4-depleted cells do not arrest in mitosis after nocodazole treatment, indicating a spindle assembly checkpoint (SAC) failure. Thus, we find that PRP4 is necessary for recruitment or maintenance of the checkpoint proteins MPS1, MAD1, and MAD2 at the kinetochores. Our data clearly identify PRP4 as a previously unrecognized kinetochore component that is necessary to establish a functional SAC.
Introduction
Chromosome segregation is a complex and dynamic process during which paired sister chromatids attached to the microtubules of the mitotic spindle are equally segregated in the two daughter cells. To avoid defects in chromosome segregation, mitotic exit is delayed until all chromosomes are properly attached to the mitotic spindle by a control mechanism named the spindle assembly checkpoint (SAC; Rieder et al., 1994; Rieder and Salmon, 1998).
SAC activation requires numerous proteins that localize at the kinetochores in early mitotic stages. The main components of this surveillance mechanism have first been identified in budding yeast and include MAD1, MAD2, MAD3 (Li and Murray, 1991), BUB 1, BUB3 (Hoyt et al., 1991), and MPS1 (Weiss and Winey, 1996). In higher organisms, all of these mitotic checkpoint proteins localize at unattached kinetochores during prometaphase. In addition, other kinetochore and centromeric proteins such as centromere protein A (CENP-A), C, I, F, E, hMIS12, the Ncd80 complex, the aurora B complex, and the RZZ (Rod, ZW10, Zwilch) complex are necessary for recruitment of the SAC and are components of the kinetochore structure (Chan et al., 2005; Vos et al., 2006). Localization of SAC proteins at kinetochores seems to be hierarchical, implying that the recruitment of some depends on the prior recruitment of others (Liu et al., 2006). In response to unattached kinetochores, the SAC is activated, resulting in the inhibition of CDC20, an activator of the multisubunit E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) that is responsible for the metaphase→anaphase transition (Kramer et al., 1998). CDC20 is sequestered together with checkpoint proteins in complexes containing MAD2, BUB3, and BUBR1, thus preventing APC/C activation and premature mitotic exit (Sudakin et al., 2001). When kinetochores are properly attached to spindle microtubules, the spindle checkpoint is turned off. This allows the APC/C to ubiquitinate mitotic proteins that will then be degraded by the proteasome machinery, triggering mitotic exit (Alexandru et al., 1999). In higher eukaryotes, SAC is required in normal conditions to check whether chromosomes are correctly attached to microtubules before anaphase onset (Li and Nicklas, 1995; Rieder et al., 1995). Inactivation of SAC genes always results in severe chromosome missegregations in mammalian cells. In addition, the depletion of SAC proteins inhibits the mitotic arrest induced by microtubule-depolymerizing drugs (Meraldi et al., 2004).
The PRP4 gene encodes a 150-kD serine-threonine protein kinase that has been implicated in the regulation of mRNA splicing in Schizosaccharomyces pombe, and mutations in PRP4 lead to the accumulation of pre-mRNAs (Rosenberg et al., 1991). However, the Drosophila melanogaster homologue protein has also turned up in screens for genes involved in mitosis (Kiger et al., 2003), and, in S. pombe, the expression of a dominant truncated PRP4 protein reportedly induced mitotic aberrations, suggesting a dual PRP4 function in RNA splicing and mitosis in this organism (Gross et al., 1997). Therefore, we decided to further explore the possible mitotic role of PRP4.
Results and discussion
We raised a polyclonal antibody against the PRP4 protein. Western blotting revealed the expected ∼150-kD protein (Fig. 1 A, left; − lane). By immunofluorescence experiments, the PRP4 protein appeared on fixed interphase cells as nuclear punctae (Fig. 2 B), which is in agreement with the localization previously described (Kojima et al., 2001; Dellaire et al., 2002). This nuclear signal disappeared in cells transfected with PRP4 siRNA (Fig. 2 B, middle; compare top with bottom) as well as the protein band by Western blotting (Fig. 1 A, + lane; and Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1).
Figure 1.
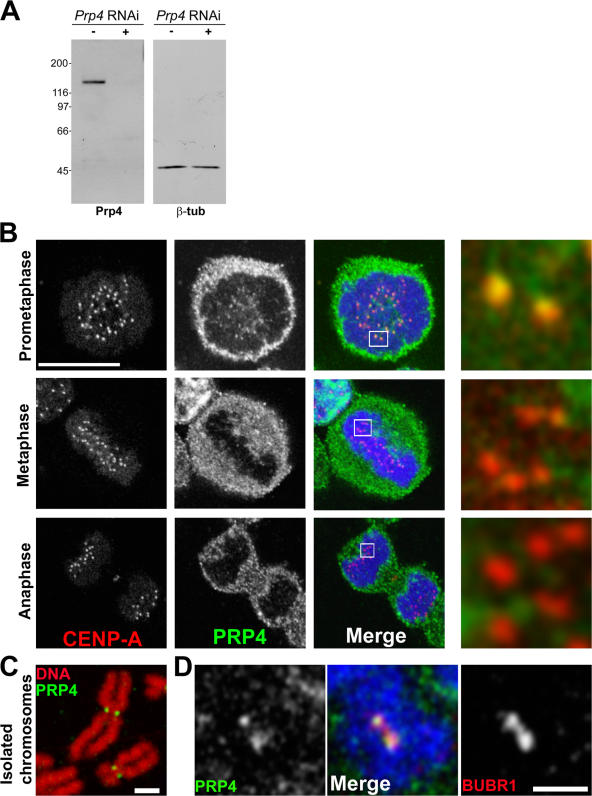
PRP4 antibody and localization during mitosis. (A) Western blot showing PRP4 (left) protein levels in control (−) and PRP4 RNAi–treated cells (+) with one of the siRNA oligonucleotides used in this study. The treatment depletes PRP4 but not β-tubulin, which was used as a loading control (right). (B) Immunolocalization of the PRP4 protein kinase in mitotic HeLa cells with affinity-purified antibodies. The cells were fixed and stained for PRP4 (green and middle panels in monocrome), CENP-A (red and left panels in monocrome), and DNA (blue). PRP4 protein displays a granular and punctiform cytoplasmic distribution that makes z stacks difficult to interpret. For this reason, only single confocal planes are shown. An enlarged view of the centromeric and kinetochore region (boxed areas) is shown on the right. PRP4 colocalization with CENP-A is mainly detected during prometaphase (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1). (C) PRP4 localization (green) was examined alone on chromosomes spreads isolated from mitotic cells (see Materials and methods). DNA is red. (D) PRP4 (green and left panel in monochrome) colocalizes with BUBR1 (red and right panel in monochrome) at the outer kinetochore region of isolated mitotic chromosomes. The chromosomes are shown in blue. Bars (B): 10 μm; (C and D) 2 μm.
Figure 2.
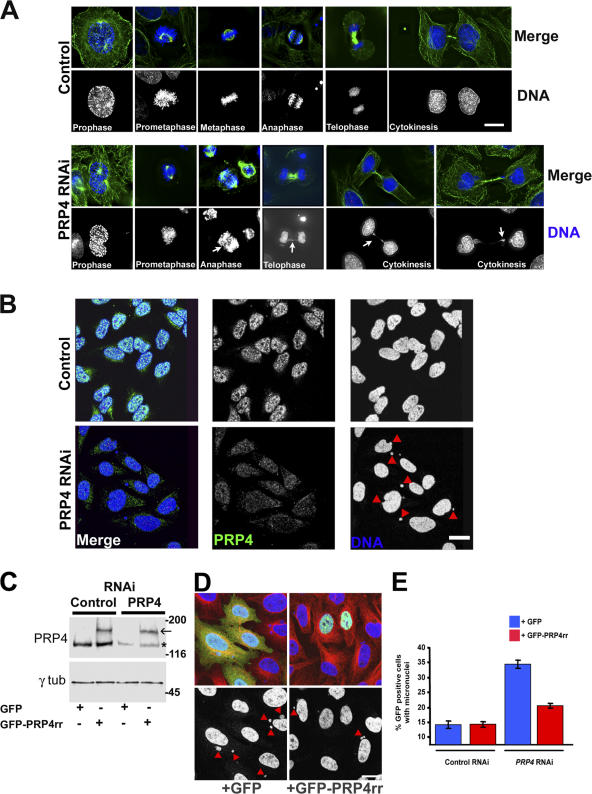
PRP4 RNAi leads to chromosome segregation defects. (A) Distribution of β-tubulin (green), γ-tubulin (red), and DNA (blue and bottom panels in monochrome) during mitosis in control (top) or PRP4 RNAi–treated cells (bottom). The mitotic stages are displayed at the bottom of each panel. 48 h after PRP4 RNAi treatment (bottom), most mitotic cells display a prometaphase-like morphology. PRP4-depleted cells exhibit lagging chromosomes during anaphase and telophase (arrows). Residual DNA bridges are often observed during cytokinesis (arrows). (B) Control (top) and PRP4-depleted cells (bottom) were stained for CENP-A (red), PRP4 (green and middle panels in monochrome), and DNA (blue and right panels in monochrome). The PRP4 knockdown cells often contain micronuclei (indicated by red triangles), whereas control nuclei are normal. (C) Control or PRP4 siRNA were cotransfected with GFP or GFP-PRP4rr (indicated at the bottom). Protein levels were analyzed by Western blotting against PRP4 (top) or γ-tubulin (bottom; as a loading control). PRP4 protein is knockdown (asterisk), whereas the GFP-PRP4 protein level remains stable (arrow) after RNAi. (D) PRP4 siRNA was cotransfected with GFP (left) or GFP-PRP4rr (right). GFP is green, microtubules are red, and DNA is blue (and monochrome in the bottom panels). The red triangles indicate the presence of micronuclei. Note the absence of micronuclei in GFP-PRP4rr–transfected cells (right), whereas the green GFP cells (left) contained numerous micronuclei. (E) Histogram showing the percentage ± SEM (error bars) of interphase GFP-expressing cells containing micronuclei. The background of the experiment is 13.0 ± 2.4% and 13.4 ± 1.9% for GFP and GFP-PRP4rr, respectively. A rescue is obtained after transfection with GFP-PRP4rr (20 ± 1.3%) but not with GFP (34.1 ± 1.8%). Bars, 10 μm.
The PRP4 protein kinase was previously shown to associate with isolated mitotic chromosome arms (Dellaire et al., 2002). To further reexamine this subcellular localization, dividing HeLa cells were methanol fixed and stained for CENP-A (as an inner kinetochore marker) and affinity-purified anti-PRP4 antibodies (Fig. 1 B). During prometaphase, most kinetochores (59.7 ± 19.1%; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1) stained positive for PRP4. During metaphase and anaphase, 27.0 ± 12.8% and 10.9 ± 7.1% of all kinetochores, respectively, displayed positive PRP4 labeling, suggesting that PRP4 leaves the kinetochore during metaphase. A strong granular signal was also detected in the cytoplasm, indicating that only a small fraction of the protein was kinetochore associated.
To confirm this kinetochore localization and eliminate the cytoplasmic PRP4 pool, isolated chromosomes prepared from mitotic cells were spread, fixed onto glass coverslips, and processed for immunofluorescence using anti-PRP4 antibodies. We found that PRP4 colocalized with BUBR1 at the outer kinetochore region (Fig. 1, C and D). Moreover, isolated chromosomes prepared from HeLa cells expressing a GFP-tagged PRP4 protein also showed a clear colocalization with MAD2 at the outer kinetochore region (Fig. S1 C).
This kinetochore localization prompted us to investigate whether PRP4 was playing a role during mitosis. To do so, we tested three siRNA oligonucleotides directed against different parts of the PRP4 mRNA. After 2 d of RNAi treatment with any of the oligo sets, PRP4 protein levels were reduced by at least 80% (Fig. S1 B) compared with control-transfected cells.
Cultures subjected to PRP4 siRNA treatment displayed reduced mitotic indexes of 3.1 (±0.8) versus 7.2% (±1.1) for control cells after 24 h and 2.8 (±0.4) versus 7.1% (±0.3) after 48 h (n = 3; ∼1,000 cells). Among the few mitotic PRP4-depleted cells, most were apparently in prometaphase and anaphase. Well-defined metaphase cells with aligned chromosomes were rare and represented only 11.9% of mitotic cells versus 46.4% in control cells (Table I). In addition, most of the anaphase and telophase cells exhibited abnormal figures (22.2 vs. 1.6% for control cells) and often showed lagging chromatids/chromosomes in the spindle midzone (Table I and Fig. 2 A). 21.0% of the PRP4-depleted cells showed residual DNA bridges during cytokinesis (Table II). As a consequence of these segregation defects, during interphase and very late cytokinesis, we observed a sevenfold increase in the frequency of cells with micronuclei (Table II and Fig. 2, A and B). We decided to check whether a GFP-tagged PRP4 RNAi-resistant construct (GFP-PRP4rr) could save the phenotype caused by PRP4 RNAi. GFP or GFP-PRP4rr were cotransfected with control or PRP4 siRNAs (Fig. 2, C–E). Western blot analyses showed that the endogenous PRP4 protein level was knocked down after RNAi, but the tagged protein remained stable (Fig. 2 C). In control RNAi conditions, GFP- or GFP-PRP4rr–positive cells contained ∼13% of micronuclei. In GFP- or GFP-PRP4rr–positive cells after PRP4 RNAi, this percentage was ∼35% and ∼20%, respectively, suggesting that the PRP4 RNA–resistant construct was rescuing the PRP4 RNAi phenotype (Fig. 2, D and E). Together, these data suggest a role for PRP4 during mitosis.
Table I. Quantification of mitotic defects after 48-h PRP4 RNAi in HeLa cells.
| RNAi treatment |
Prophase | Prometaphase | Metaphase | Anaphase | Telophase | Lagging chromatids during anaphase/telophase |
|---|---|---|---|---|---|---|
| % | % | % | % | % | % | |
| Control RNAi | 7.8 ± 1.5 | 28.4 ± 4.6 | 46.4 ± 3.6 | 8.8 ± 2.4 | 7.0 ± 1.4 | 1.6 ± 1.5 |
| PRP4 RNAi | 9.7 ± 5.1 | 42.9 ± 7.5 | 11.9 ± 13.2 | 6.0 ± 2.7 | 7.3 ± 6.3 | 22.2 ± 11.6 |
Quantification of mitotic defects after 48-h PRP4 RNAi in HeLa cells. Distribution of mitotic cells in the different stages of mitosis: prophase, prometaphase, metaphase, anaphase, and telophase. The values indicate the percentage of cells in each phase ± SD. A supplemental class has been added for aberrant mitosis. PRP4-depleted cells exhibited abnormal anaphase and telophase figures with lagging chromosomes. Note that the mitotic index is two times lower in PRP4-depleted cells.
Table II. Quantification of aberrant cells during cytokinesis and interphase after 48-h PRP4 RNAi in HeLa cells.
| RNAi treatment | Cytokinesis
|
Interphase cells
|
|
|---|---|---|---|
| With micronuclei | With DNA bridge | With micronuclei | |
| % | % | % | |
| Control RNAi | 6.2 ± 1.2 | 3.0 ± 2.9 | 3.9 ± 0.8 |
| PRP4 RNAi | 44.2 ± 13.9 | 21.0 ± 5.56 | 30.9 ± 6.4 |
Quantification of aberrant cells in cytokinesis and interphase after PRP4 RNAi. The values indicate the percentage of cytokinesis or interphase cells displaying residual DNA bridges and/or micronuclei after PRP4 RNAi treatment.
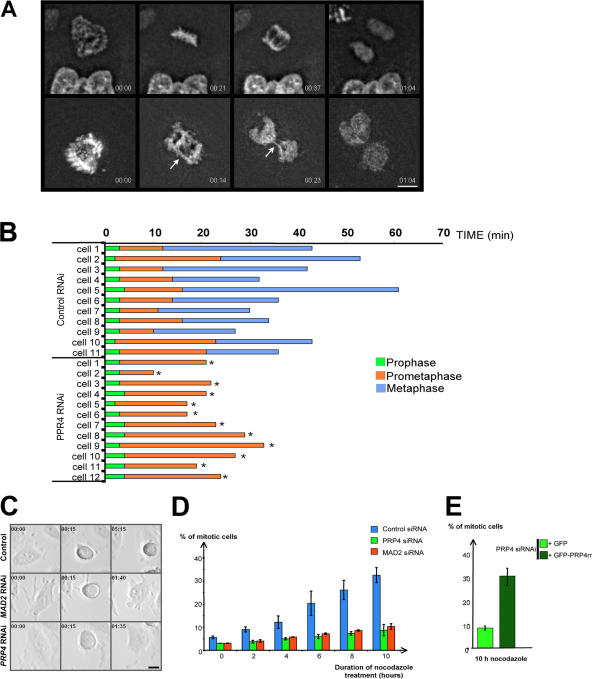
To further characterize chromosome dynamics and the timing of mitosis in control and PRP4-depleted cells, we filmed HeLa cells expressing an H2B-GFP transgene (Kimura and Cook, 2001). In control HeLa H2B-GFP cells (n = 11), chromosomes correctly aligned to form a metaphase plate (Fig. 3 A, top; panel at 21 s) before anaphase onset (Fig. 3 A, top; panel at 37 s [control]; and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1). After PRP4 RNAi (n = 12), anaphase always started before chromosome alignment (Fig. 3 A, bottom; panel at 23 s; and Video 2). Fig. 3 B also shows that all recorded PRP4-depleted cells do not congress their chromosomes to the metaphase plate and that mitotic exit occurs earlier than control cells. Together, these data show that PRP4 is required for chromosome congression and segregation but also for mitotic timing.
Figure 3.
PRP4 depletion prevents correct chromosome congression and segregation and induces premature anaphase onset and SAC override. (A) HeLa cells expressing H2B-GFP were filmed at the onset of mitosis to monitor chromosome dynamics in control (n = 11; top) or PRP4 siRNA–transfected cells (n = 12; bottom). Time in hours/minutes is displayed at the bottom right of each panel. The metaphase plate is formed 21 min after NEBD, and anaphase onset starts after 37 min in the control (A, top; also see Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1). After PRP4 RNAi, anaphase starts 14 min after NEBD, although all chromosomes have not aligned on a metaphase plate, leading to lagging chromosomes. This results in a chromatin bridge (23 s and arrows, bottom; also see Video 2). (B) Prophase (green), prometaphase (orange), and metaphase duration (blue) are displayed for each control (top) or PRP4 RNAi–treated cell (bottom). The asterisks are indicative of anaphase onset before metaphase plate formation. The histogram shows that PRP4-depleted cells fail to congress their chromosomes to the metaphase plate at a time when this event is achieved in the control cells. (C) HeLa cells were treated with nocodazole 1 h before filming to trigger microtubule depolymerization and SAC activation. The figure represents selected frames of nocodazole-treated HeLa cells transfected with control (top), MAD2 (middle), or PRP4 siRNA (bottom). In control (top), the cell enters mitosis (as judged by cell rounding; 15 s) and remains blocked at this stage for the duration of the observation (5 min 15 s), indicating that the SAC is activated. The MAD2- (middle) and PRP4 (bottom)-depleted cells enter mitosis (15 s) but return into interphase, as observed by the cell reflattening (1 min 40 s and 1 min 35 s, respectively), indicating a SAC override. The time is displayed (hours/minutes) at the top left of each panel. (D) Control (blue), MAD2- (red), or PRP4 siRNA–treated cells (green) were incubated in the presence of nocodazole 24 h after siRNA transfection, and the mitotic indices were analyzed at 0, 2, 4, 6, 8, and 10 h. The values show the mean mitotic indices ± SEM (error bars). Control cells arrest normally in mitosis, unlike MAD2- and PRP4-depleted cells, indicating a SAC failure. (E) The PRP4-depleted cells were also cotransfected with GFP or GFP-PRP4rr constructs and treated with nocodazole for 10 h. Although depleted cells expressing GFP still show a reduced mitotic index, a full rescue is observed in the presence of the GFP-PRP4rr protein. Bars, 10 μm.
To confirm this acceleration, the time between nuclear envelope breakdown (NEBD) and anaphase onset was analyzed by live cell imaging of HeLa cells. This time was 56.2 ± 23.1 min (n = 36) in control cells (Fig. S2 B, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1). In PRP4-depleted cells, this time was only 43.1 ± 12.9 (n = 43), 46.4 ± 7.8 (n = 10), and 33.6 ± 8.0 min (n = 36) after transfection with the different PRP4 siRNAs (Fig. S2 B). The mitosis peak time (Meraldi et al., 2004) was 49 min for the control cells and 38, 41, and 31 min for PRP4 knockdown cells (siRNAs 1–3), confirming that mitosis duration was reduced after PRP4 RNAi (Fig. S2 A).
The S. pombe PRP4 gene was previously shown to be required for pre-mRNA splicing (Rosenberg et al., 1991; Gross et al., 1997). We then checked whether this process was compromised after PRP4 RNAi in HeLa cells (see Materials and methods and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1). Pre-mRNA splicing was identical to control cells. Thus, the mitotic effect of PRP4 knockdown is unlikely to be a secondary consequence of general splicing defects.
These defects were similar to those previously observed after the depletion of other SAC components such as MAD2 and BUBR1: shorter mitosis duration and chromosome segregation defects (Meraldi et al., 2004). In addition, MAD2- and BUBR1-depleted cells do not arrest in mitosis after treatment with nocodazole, a drug that depolymerizes mitotic spindle microtubules.
To test for a possible role of PRP4 in SAC signaling, we monitored mitotic progression by time-lapse video microscopy in the presence of 50 ng/ml nocodazole, conditions in which SAC is activated. When control cells entered mitosis, they rounded and remained blocked in that stage for the duration of the experiment (∼5 h; n > 25), indicating a mitotic arrest caused by the presence of a functional SAC (Fig. 3 C, top). In contrast, when MAD2- or PRP4-depleted cells were challenged in the same conditions, the cells rounded and became adherent after 102.5 ± 14.9 min (n = 9) and 97.0 ± 12.3 min (n = 10), respectively, indicating their return into interphase without chromosome segregation (Fig. 3 C, middle and bottom). In addition, the mitotic index was analyzed in a time course experiment in control, MAD2-, or PRP4-depleted cells in the presence of nocodazole (Fig. 3 D). Although the mitotic index increases during the experiment in control cells, indicating checkpoint activation and mitotic arrest, the mitotic index did not increase in MAD2- and PRP4-depleted cells, indicating a SAC failure. In parallel, a rescue experiment was performed by coexpression of the PRP4 siRNA with GFP or GFP-PRP4rr constructs described previously (Fig. 3 E). The mitotic index of PRP4-depleted cells was low in the presence of GFP. In contrast, a full rescue was observed in the presence of the GFP-PRP4rr construct, indicating that the GFP-tagged PRP4 protein was able to complement PRP4 depletion.
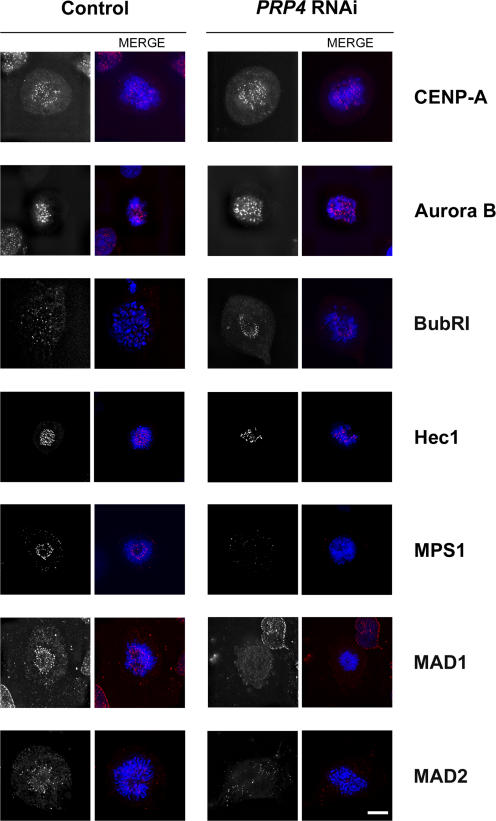
To investigate how the absence of the PRP4 protein affected the SAC, we examined whether the PRP4 RNAi treatment disturbed the kinetochore localization of several proteins involved in the SAC. The CENP-A, aurora B, BUBR1, and Hec1 proteins were all localized at the kinetochores of prometaphase PRP4-depleted cells (Fig. 4; Liu et al., 2006). On the other hand, MPS1, MAD1, and MAD2 were not detected at these kinetochores, although their protein levels were normal by Western blotting (Figs. 4 and S1 A). Therefore, our data indicate that PRP4 is specifically required for the ordered recruitment/maintenance of MPS1, MAD1, and MAD2 at the kinetochore.
Figure 4.
PRP4 knockdown prevents MPS1, MAD1, and MAD2 recruitment to the kinetochores. Localization of centromeric and kinetochore proteins on control (left) or PRP4-depleted cells (right) during prometaphase. Fixed cells are stained from top to bottom for CENP-A, aurora B, BUBR1, Hec1, MPS1, MAD1, and MAD2 (shown in monochrome and in red in the merge panels) and are costained for DNA (blue). CENP-A, aurora B, BUBR1, and Hec1 show a normal localization, whereas MPS1, MAD1, and MAD2 were absent after PRP4 depletion. Bar, 10 μm.
The fission yeast PRP4 homologue gene seems to be necessary for pre-mRNA splicing (Gross et al., 1997), but our experiments show that this function is not conserved or required for cell viability in the human (Fig. S3). Moreover, all of the checkpoint proteins analyzed so far by Western blotting proved to be expressed at control levels. This indicates that the defects observed in this study are direct and are not caused by the disappearance of known checkpoint proteins. In agreement with a direct function for PRP4 during the checkpoint, a pool of the PRP4 protein localizes at the kinetochores of isolated chromosomes, the right place to perform this mitotic function. Moreover, PRP4 has recently been identified as a component of the human mitotic spindle phosphoproteome, which contains kinetochore-associated proteins (Nousiainen et al., 2006).
In this study, we found that PRP4 is important for chromosome alignment. In addition, several lines of evidence support our conclusion that PRP4 belongs to the family of SAC regulatory genes. First, after PRP4 depletion, severe chromosome segregation defects are observed. Second, the cells do not arrest in mitosis in the presence of nocodazole. By themselves, these two criteria indicate a requirement of PRP4 for SAC function. This effect can be easily explained by the lack of MPS1 recruitment that is itself needed for the recruitment of MAD1 and MAD2.
Interestingly, PRP4 also reduces the mitotic duration. This places PRP4 in the checkpoint category, including MAD2 and BUBR1 (components of the soluble mitotic checkpoint complex), which are required to maintain a minimal mitosis duration (May and Hardwick, 2006). How does PRP4 mediate this additional function? The simplest hypothesis could be that PRP4 regulates the cytoplasmic pool of the mitotic checkpoint complex. Indeed, even if a fraction of the PRP4 protein is localized at the kinetochore region, most of the protein is cytoplasmic with a granular and punctiform distribution during mitosis. This localization is intriguing compared with those of previously characterized checkpoint proteins. Regardless, PRP4 appears to be an important gene regulating SAC, and further studies will be required to provide additional mechanistic insights into how PRP4 controls checkpoint function and mitotic duration.
Materials and methods
Cell culture
HeLa and HeLa stably expressing GFP-tagged H2B (provided by Dr. H. Kimura, University Of Kyoto, Kyoto, Japan) cell lines were maintained in DME supplemented with 10% FCS and antibiotics (penicillin and streptomycin).
Interference RNA
siRNA oligonucleotides were purchased from Eurogentec. For human PRP4 RNAi, three different sequences were used: 5′-UGCAAGAGCCAACCAAGAA-3′, 5′-GCAGGAAUCUUCGUCUGAU-3′, and 5′-UGAUAUGUUUGCUGCGUAU-3′. For the rescue experiment, a siRNA with the sequence 5′-AGACCAACGUAAGAAAGUA-3′ was used. For MAD2 RNAi, the sequence used was 5′-UACGGACUCACCUUGCUUG-3′ (Tang et al., 2004). A random siRNA was used as a control. siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Each experiment was repeated at least three times.
Plasmid constructions
The PRP4 siRNA-resistant construct (GFP-PRP4rr) was generated by PCR using the vector pBlueScriptR-PRP4 (IMAGE clone 4821304; GenBank/EMBL/DDBJ accession no. BC034969) and two primer pairs (mutations and restriction sites are underlined) with the following sequences: (A) 5′-TCAGAATTCCCGCCGCGGAGACCCAG-3′, (B) 5′-CCTTTAGCTGGTGTACCTTTTTGCGCTGATCTTCAGG-3′, (C) 5′-CCTGAAGATCAGCGCAAAAAGGTACACCAGCTAAAGG-3′, and (D) 5′-CTTGGATCCGTTTAAATTTTTTCCTGGATG-3′. Two PCRs were performed with the oligonucleotides A/B and C/D. Both PCR products were mixed and amplified with primers A and D. The resulting fragment was cloned into pEGPC3 (BD Biosciences) using EcoRI and BamHI restriction sites to generate the GFP-PRP4rr construct. The mutations were confirmed by DNA sequencing. To generate a recombinant histidine-tagged PRP4(aa 504–688), a partial sequence of human PRP4 was amplified by PCR from the vector pBlueScriptR-PRP4 using the primers 5′-GAAGAATTCTTTAAAGGAAGTCTTTCTGAA-3′ and 5′-TTGCGGCCGCATTGTAACGTTTATCTAGGA-3′ (restriction sites are underlined). The PCR product was cloned into the pET-21a expression vector (EMD) using EcoRI and NotI restriction sites.
Production of a recombinant protein PRP4(aa 504–688)-His and anti-PRP4 antibody
Competent Escherichia coli BL21 was transformed with pET-21a-PRP4(aa 508–688) (EMD). The recombinant protein expression was induced with 1 mM IPTG in cells during 4 h at 25°C. The protein was purified according to the manufacturer's instructions (QIAGEN), dialyzed overnight in PBS at 4°C, and used for rabbit immunization.
Live cell imaging and drug treatment
To monitor chromosome dynamics, HeLa cells carrying GFP-tagged H2B were observed 48 h after RNAi treatment. We used an inverted fluorescence video microscope (DMIRB; Leica) with a 63× NA 1.40 objective lens equipped with a Xenon lamp (time exposure of 4 ms) and camera (CoolSNAP HQ; Roper Scientific). Z series (six planes, 1-μm step, bin × 2) were acquired every 1 min.
To investigate the duration of mitosis, HeLa cells transfected with control or with each PRP4 siRNA oligonucleotide were filmed on an inverted video microscope (time exposure of 10 ms) for 48 h after RNAi treatment. Images were acquired every 1 min. Videos were analyzed to determine the delay between the NEBD and anaphase onset.
For SAC functional test, control or PRP4-depleted cells were incubated with 50 ng/ml nocodazole 1 h before filming on an inverted video microscope with a 20× NA 0.40 objective lens (time exposure of 10 ms). One image was acquired every 5 min. Alternatively, control, MAD2, or PRP4 siRNA–treated cells were incubated in the presence of nocodazole, and the mitotic indices were measured at different time points (0–10 h). Each experiment was repeated at least three times for each siRNA (1–3). Images were treated with MetaMorph software (MDS Analytic Technologies).
Immunofluorescence analyses on fixed cells
HeLa cells were grown on glass coverslips and fixed using three different protocols. To observe the phenotype after RNAi treatments, cells were fixed for 10 min in 75% methanol, 3.7% formaldehyde, and 0.5× PBS, permeabilized for 2 min in PBS containing 0.1% Triton X-100, and washed three times during 5 min in PBS.
For kinetochore and centromeric protein localization, cells were fixed for 30 min in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 8 mM MgCl2) containing 4% PFA and 0.5% Triton X-100 and were washed three times during 5 min in PBS. To monitor PRP4 protein localization at the kinetochores, the cells were prepermeabilized for 60 s in PBS and 0.1% Triton X-100 and fixed for 10 min at −20°C in methanol.
Cells on coverslips were blocked for 1 h in PBS containing 0.1% Triton X-100 and 1% BSA (PBSTB) and were incubated for 2 h at RT with the primary antibodies in PBSTB. Secondary antibodies were incubated for 1 h at RT. Between each incubation, coverslips were washed three times for 5 min with PBSTB. Coverslips were mounted in Vectashield containing DAPI (Vector Laboratories) to stain the DNA. Samples were viewed with a fluorescent microscope (DMRXA2; Leica) with a 63× NA 1.32 lens equipped with standard fluorescence filters. The images were acquired with a CoolSNAP ES camera using MetaMorph software and prepared as single sections or maximum intensity projections before being processed in Photoshop 7.0 (version 7.0; Adobe).
Immunofluorescence on purified chromosomes
500 ng/ml nocodazole was added to cultured cells for 2 h. Mitotic control cells or cells expressing the GFP-PRP4rr construct were harvested by mitotic shake. Culture medium with mitotic cells was recovered and supplemented with 75 mM KCl for 30 min at 37°C. Cells were spun onto coverslips at 2,000 rpm for 10 min. Coverslips were fixed for 15 min in PBS containing 4% PFA and were rinsed in PBS and processed for immunofluorescence. Staining was viewed with an inverted confocal microscope (DM IRE2 SP2; Leica) equipped with a 63× NA 1.4 objective lens using the LCS 3D software (Leica).
Antibodies and Western blotting
In this study, we used monoclonal YL1/2 rat antityrosinated tubulin (Chemicon), polyclonal rabbit anti-MAD2, mouse monoclonal anti-HA (Covance), monoclonal mouse anti-HEC1 (GeneTex), monoclonal mouse anti-MPS1 (Invitrogen), monoclonal mouse anti–CENP-A and anti-MAD1 (Abcam), monoclonal mouse anti-BUBR1 and anti–aurora B (BD Biosciences), monoclonal GTU-88 mouse anti–γ-tubulin and mouse anti–β-tubulin (clone 2.1) antibodies (Sigma-Aldrich), and mouse monoclonal anti-MAD2 antibody (Institute of Molecular Oncology Foundation). Anti-PRP4 rabbit affinity-purified antibodies were used at 1 μg/ml. Secondary FITC, TRITC, and peroxydase-conjugated antibodies were purchased from Jackson ImmunoResearch Laboratories.
Western blots were performed according to standard procedures, and proteins were visualized using the ECL chemiluminescent reagent (Thermo Fisher Scientific). To quantify Western blot protein levels, phosphatase-conjugated secondary antibodies were used (Jackson ImmunoResearch Laboratories) and visualized with the Enhanced ChemiFluorescence reagent (GE Healthcare). Quantification of PRP4 fluorescence signal on the nitrocellulose membrane was performed using the Storm840 apparatus (Molecular Dynamics) with ImageQuant software (GE Healthcare) and β-tubulin as a loading control.
Splicing assay
Cells were transfected with the plasmid pAdCMV-glob (Estmer Nilsson et al., 2001) and/or siRNA using Lipofectamine 2000. After 48 h, cells were collected, and total RNA was isolated with the Nucleospin RNA II kit (Macherey Nagel) and treated with 4 U Turbo DNase (Ambion). Reverse transcription reactions were performed in the presence of 1 μg of total RNA, 0.5 μg oligo dT(12–18), and 200 U SuperScript II Reverse Transcriptase (Invitrogen) for 50 min at 42°C. PCR reactions were performed with the Expand high-fidelity PCR system (Roche) for 40 cycles. Primers were complementary to exons 1 and 2 of the β-globin plasmid but not for the endogenous β-globin gene. PCR products were separated in agarose gels. If transgenic mRNA splicing is effective, a 334-bp PCR product is generated. In case of defective mRNA processing, a longer 464-bp product is generated.
Online supplemental material
Fig. S1 shows protein levels in control and PRP4-depleted cells and GFP-PRP4 kinetochore localization. Fig. S2 presents quantification of mitosis duration in control and PRP4-depleted cells. Fig. S3 shows that human PRP4 knockdown does not inhibit RNA splicing. Video 1 shows a control mitotic HeLa cell expressing GFP-tagged H2B 48 h after control RNAi. Video 2 shows a mitotic HeLa cell expressing GFP-tagged H2B 48 h after PRP4 RNAi. Table S1 shows the percentages of PRP4-labeled kinetochores during prometaphase, metaphase, and anaphase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200703133/DC1.
Supplemental Material
Acknowledgments
We wish to thank Dr. Hiroshi Kimura for the H2B GFP-tagged HeLa cell line as well as Dr. Legagneux, Dr. Mereau, and Dr. Hardy for their technical assistance. We also thank Emeric Sevin for English revision and laboratory members for critical readings. Special thanks are also due to Roger Karess for useful advice, critical reading, and support.
This work was supported by the Ligue Nationale Contre le Cancer Equipe Labellisée and Cancéropôle Grand Ouest. E. Montembault is a fellow of the French Ministère de la Recherche.
Abbreviations used in this paper: APC/C, anaphase-promoting complex/ cyclosome; CENP-A, centromere protein A; NEBD, nuclear envelope breakdown; SAC, spindle assembly checkpoint.
References
- Alexandru, G., W. Zachariae, A. Schleiffer, and K. Nasmyth. 1999. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 18:2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G.K., S.T. Liu, and T.J. Yen. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598. [DOI] [PubMed] [Google Scholar]
- Dellaire, G., E.M. Makarov, J.J. Cowger, D. Longman, H.G. Sutherland, R. Luhrmann, J. Torchia, and W.A. Bickmore. 2002. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22:5141–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estmer Nilsson, C., S. Petersen-Mahrt, C. Durot, R. Shtrichman, A.R. Krainer, T. Kleinberger, and G. Akusjarvi. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, T., M. Lutzelberger, H. Weigmann, A. Klingenhoff, S. Shenoy, and N.F. Kaufer. 1997. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 25:1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., L. Totis, and B.T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517. [DOI] [PubMed] [Google Scholar]
- Kiger, A.A., B. Baum, S. Jones, M.R. Jones, A. Coulson, C. Echeverri, and N. Perrimon. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., and P.R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, T., T. Zama, K. Wada, H. Onogi, and M. Hagiwara. 2001. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 276:32247–32256. [DOI] [PubMed] [Google Scholar]
- Kramer, E.R., C. Gieffers, G. Holzl, M. Hengstschlager, and J.M. Peters. 1998. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol. 8:1207–1210. [DOI] [PubMed] [Google Scholar]
- Li, R., and A.W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531. [DOI] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., J.B. Rattner, S.A. Jablonski, and T.J. Yen. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, K.M., and K.G. Hardwick. 2006. The spindle checkpoint. J. Cell Sci. 119:4139–4142. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., V.M. Draviam, and P.K. Sorger. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. [DOI] [PubMed] [Google Scholar]
- Nousiainen, M., H.H. Sillje, G. Sauer, E.A. Nigg, and R. Korner. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA. 103:5391–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and E.D. Salmon. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., A. Schultz, R. Cole, and G. Sluder. 1994. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., R.W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, G.H., S.K. Alahari, and N.F. Kaufer. 1991. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. 226:305–309. [DOI] [PubMed] [Google Scholar]
- Sudakin, V., G.K. Chan, and T.J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., Y. Sun, S.E. Harley, H. Zou, and H. Yu. 2004. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 101:18012–18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, L.J., J.K. Famulski, and G.K. Chan. 2006. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem. Cell Biol. 84:619–639. [DOI] [PubMed] [Google Scholar]
- Weiss, E., and M. Winey. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.