Abstract
Along-standing biological question is how a eukaryotic cell controls the size of its nucleus. We report here that in fission yeast, nuclear size is proportional to cell size over a 35-fold range, and use mutants to show that a 16-fold change in nuclear DNA content does not influence the relative size of the nucleus. Multi-nucleated cells with unevenly distributed nuclei reveal that nuclei surrounded by a greater volume of cytoplasm grow more rapidly. During interphase of the cell cycle nuclear growth is proportional to cell growth, and during mitosis there is a rapid expansion of the nuclear envelope. When the nuclear/cell (N/C) volume ratio is increased by centrifugation or genetic manipulation, nuclear growth is arrested while the cell continues to grow; in contrast, low N/C ratios are rapidly corrected by nuclear growth. We propose that there is a general cellular control linking nuclear growth to cell size.
Introduction
The sizes of membrane-bound organelles are important for eukaryotic cell organization and function. Although progress has been made in understanding the biochemical mechanisms underlying how membranes are formed and shaped, little is known about how the overall sizes of these organelles are determined (Nurse, 1975, 1985; Marshall, 2004; Umen, 2005; Zimmerberg and Kozlov, 2006). The nucleus is a good organelle to study this problem because it is usually present as a single organelle in each cell and is easy to visualize accurately. Despite over 100 years of research it remains poorly understood how nuclear size is regulated. In 1903 Richard Hertwig formulated the first quantitative hypothesis about organelle size (Hertwig, 1903). Based on work on sea urchin embryos and algae he proposed the “Kern-Plasma-Relation” or karyoplasmic ratio, as a constant characteristic for every cell type (Gerassimow, 1902; Boveri, 1905). Positive correlations among nuclear size, cell size, genome size, ploidy, or generation times have been described, often comparing a diverse range of species, although mechanistic explanations have remained elusive (Schmidt and Schibler, 1995; Gregory, 2005; Jovtchev et al., 2006). One idea for nuclear size control is the nucleoskeletal theory, which states that nuclear volume is determined by the amount of DNA, the degree of DNA compaction, the amount and composition of nuclear membranes, and in metazoa the components of the nuclear lamina (Orgel and Crick, 1980; Cavalier-Smith, 1982; Gregory, 2005). However, it does not explain well how large changes in cell size and nuclear size can occur between cells within a single organism, often linked to differentiation or developmental programs of the cell (Sato et al., 1994; Taddei et al., 2004; Brandt et al., 2006). In contrast to metazoan cells, yeasts do not reassemble the nuclear envelope (NE) after mitosis and thus provide simple models to study the regulation of nuclear size in a single cell. Recent work in budding yeast has shown that nuclear and cell size correlate in cell size mutants as well as throughout the cell cycle and in diploid cells (Jorgensen et al., 2007). Here we report a detailed analysis of the coordination between nuclear and cell growth in the fission yeast Schizosaccharomyces pombe. This work demonstrates a remarkably tight link between nuclear and cell size throughout a wide variety of cell sizes and physiological conditions, and suggests there is a general cellular control linking nuclear growth to cell size.
Results and discussion
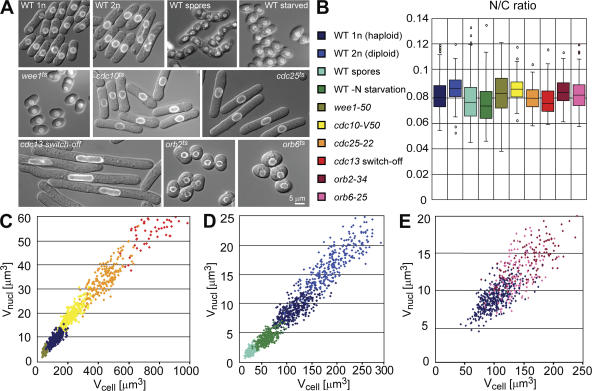
We measured nuclear and cellular volumes in individual cells of the fission yeast S. pombe using a nuclear membrane marker Cut11-GFP and Nomarski optics (West et al., 1998). In vegetatively growing wild-type cells, the ratio of nuclear to cell volume (N/C ratio) was found to be 0.080 ± 0.013 (Fig. 1, A and B), in agreement with measurements from electron microscopic tomography (Höög et al., 2007). As in budding yeast, there is a linear correlation between cell size and nuclear size (r = 0.68; Jorgensen et al., 2007). The N/C ratio and the linear correlation were unchanged in two round mutants, indicating that cell volume and not cell length correlates with nuclear size (orb2ts and orb6ts; Fig. 1, A, B, and E; Verde et al., 1995). To determine if the N/C ratio remained constant in cells of different sizes, we measured nuclear and cell volumes in spores, small cells (wee1ts), large cells (using the cell cycle mutants cdc25ts, cdc10ts, and a cdc13 switch-off strain), and wild-type cells (Nurse et al., 1976; Nurse, 1985; Russell and Nurse, 1986; Hayles et al., 1994). We found a very strong positive correlation between nuclear and cell volume (r = 0.97, n = 2136, for correlation coefficients (r) of individual distributions; see Fig. 1). Despite a 35-fold difference in average cell size, the N/C ratio was remarkably constant, varying only between 0.076 ± 0.013 and 0.089 ± 0.017 (Fig. 1, A–C). Taking the extremes of single cells, the actual span in cell size between the smallest spore and the largest cdc13 switch-off cell was 160 fold. Even cells of different sizes generated by nitrogen starvation or changes in ploidy had broadly similar N/C ratios (Fig. 1, A, B, and D). To investigate if disruption of the cytoskeleton has effects on nuclear size, we treated cells with Latrunculin A to depolymerize actin patches and cables and with MCB, which depolymerizes microtubules. These treatments did not cause obvious changes in the N/C ratio (unpublished data). During the course of those experiments we discovered that in vegetatively growing cells the volume of the nucleolus remained in a similar proportion to the nucleus (0.24 ± 0.06, n = 31, r = 0.77), despite the wide range of cell sizes (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1). We conclude that nuclear and nucleolar sizes are strongly coordinated with cell size.
Figure 1.
Nuclear and cell size correlate over a large range in cell size and shape. Quantitation of nuclear and cell size in cell size and cell shape mutants. (A) Overlay image of cell (differential interference contrast; DIC) and nuclear envelope (cut11-GFP). (B) Box-and-Whisker plots showing the N/C ratio. (C–E) Scatter plots of cell size and nuclear size. Wild type (WT): haploid (25°C, n = 382, r = 0.72), spores (25°C, n = 329. r = 0.63), nitrogen-starved cells (12 h EMM-N, 25°C, n = 275, r = 0.64), diploid (25°C, n = 308, r = 0.79). Cell size mutants: wee1-50, temperature-sensitive (ts) allele of the mitotic inhibitor wee1 resulting in a short G2 phase and mitosis at a smaller cell size (6 h 36°C, n = 179, r = 0.70), cdc10-V50, ts allele of a transcription factor controlling G1-S specific gene expression leading to a cell cycle arrest in G1 phase (3 h 36.5°C, n = 322, r = 0.82), cdc25-22, ts allele of the cdc25 phosphatase, an inducer of mitosis which arrests cells in G2 phase of the cell cycle (3 h 36.5°C, n = 188, r = 0.81), pnmt1-cdc13, a repressible allele of the b-type cyclin which leads to consecutive rounds of over-replication or arrest in G2 phase of the cell cycle (10 h release, n = 98, r = 0.62), orb2-34, a ts allele of PAK-related polarity kinase orb2 (5h 36.5°C, n = 122, r = 0.66), orb6-25, a ts allele of a kinase which coordinates cell morphogenesis with the cell cycle (5.5 h 36.5°C, n = 129, r = 0.69).
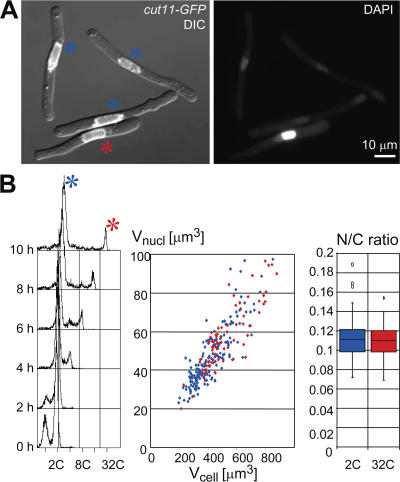
We next tested N/C ratios in fission yeast cell cycle mutants that were arrested in G1 or G2 phase with either 1C or 2C DNA content, and in diploid cells, which have twice the DNA content of the haploid cells. Examination of the plots (Fig. 1, B and D) demonstrated that cells of similar size but containing nuclei with different DNA contents had similar N/C ratios. More dramatically, the cdc13 switch-off strain produced some cells with a single nucleus of a 2C DNA content and others with a 32C DNA content as determined by FACS and DAPI staining (Fig. 2, A and B; Hayles et al., 1994). Despite the 16-fold difference in DNA content, the relative nuclear sizes were the same and N/C ratios were indistinguishable (Fig. 2, C and D). We conclude that in growing cells, the DNA content of a nucleus has no direct effect on nuclear size. Rather, ploidy determines cell size at division and in turn the sizes of the subsequent daughter cells determine the sizes of their nuclei. This result is consistent with recent work examining haploid and diploid budding yeast in both G1 and G2 phase (Winey et al., 1997; Jorgensen et al., 2007).
Figure 2.
DNA content does not directly determine nuclear size. Quantitation of nuclear and cell size in over-replicating cells. pnmt1-driven switch-off of the cyclin Cdc13 induces over-replication in a subset of cells (marked in red). Cells arrested with 2C DNA content are marked in blue. (A) For quantitative DAPI staining (right panel), cells were formaldehyde fixed 10 h after cdc13 switch-off. Left panel: overlay of DIC and Cut11-GFP. Bar, 10 μm. (B) FACS profile for DNA content shows the kinetics of cdc13 switch-off dependent over-replication (log scale FL2-H). Cell and nuclear size are represented in a scatter plot, N/C ratio in a Box-and-Whisker plot.
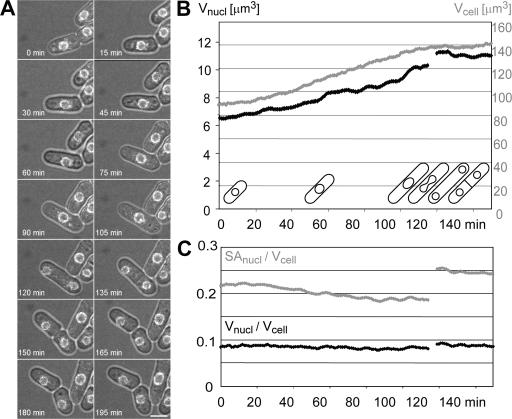
If overall cell size determines nuclear size, then the gradual increase in cell size during the cell cycle should be linked to a gradual increase in nuclear size. To test this prediction we monitored the N/C ratio using time-lapse microscopy of living wild-type cells proceeding through the cell cycle (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1). As expected, nuclear and cell volume increased concurrently, maintaining an almost constant N/C ratio throughout the cell cycle (Fig. 3). Fission yeast undergoes a rapid mitosis, with no breakdown of the NE. Measurements of nuclear volume and surface area just before and immediately after mitosis established that the combined volumes of the two daughter nuclei were approximately equal to the volume of the undivided parental nucleus. In contrast, the combined surface areas of the two daughter nuclei were much increased (Fig. 3 C). These results indicate that rapid expansion of the NE during mitosis is necessary to maintain the N/C ratio the same in the two daughter cells. A more flexible ruffled NE was observed in cells undergoing closed mitoses, which could contribute to the rapid expansion of the NE (unpublished data). Consistently, an expansion of the NE has also been reported before and after mitosis in budding yeast, where equal amounts of DNA are partitioned into two nuclei of different size (Gasser, 2002).
Figure 3.
N/C ratio is constant throughout cell cycle. Cell cycle analysis of nuclear and cell size using time-lapse microscopy (6 z-sections/min). (A) Time points of a selected field from Video 1. (B and C) The graphs represent the median of a 10 min moving averages from 5 independent cells, analyzed for an entire cell cycle as shown by the cartoons. Average cell volume (gray) and nuclear volume (black) cells and the respective N/C ratios (gray: nuclear surface area/cell volume, black: nuclear volume/cell volume). Video 1 is available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1.
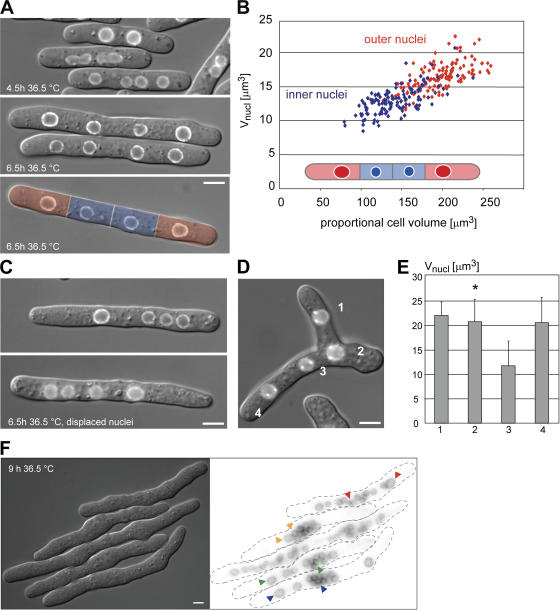
The experiments described above indicate that nuclear size is proportional to cell size. We next generated multinucleated cells using a cytokinesis mutant (cdc11ts; Nurse et al., 1976) to test two contrasting models regarding the role of nuclear position in growth control. First, nuclear growth may depend on nuclear position and the proportional amount of cytoplasm surrounding a nucleus; second, nuclear growth may be similar throughout the cell and independent of nuclear position. After two mitoses at the restrictive temperature, cells contain four nuclei of equal size that are unevenly distributed within the cell, allowing their direct comparison in different subcellular positions. Each nucleus was attributed a proportional cytoplasmic volume, defined by the midpoint between neighboring nuclei and/or cell ends. We found that more closely spaced inner nuclei grew slower than the outer nuclei that are surrounded by a larger proportional cytoplasmic volume. During interphase, the proportional N/C ratio of single nuclei was gradually adjusted until they reached a size close to the proportional cytoplasmic volume (Fig. 4 A). We propose that the local cytoplasmic environment is an important determinant of nuclear size, and further tested this hypothesis in three experiments. To distinguish between effects linked to the proportional cytoplasmic volume and the proximity to an actively growing cell tip, we first artificially displaced nuclei by centrifugation altering the distribution of nuclei within the cell. Interestingly, the nucleus that was surrounded by a larger cytoplasmic sub-volume grew faster than the others (Fig. 4 C). Second, we confirmed the hypothesis using a branching mutant, which creates an additional growth zone at the middle of a cell (tea1Δ; Snell and Nurse, 1994). In agreement with our proposal, nuclei residing in a larger cytoplasmic domain became even larger, and previously small nuclei that were close to an additional growth zone increased in size (Fig. 4, D and E). Lastly, we observed that in cdc11ts cells containing 16 or 32 nuclei, the sizes of the nuclei varied up to 10-fold depending on the amount of cytoplasm in their vicinity (Fig. 4 F). This result is also consistent with the cytoplasmic volume determining nuclear size.
Figure 4.
The proportional cytoplasmic volume directly influences nuclear size. (A) Multiple unevenly distributed nuclei within a cell are created using a cdc11-119 arrest for 4.5 to 6.5 h. During this interval, nuclear size of the outer nuclei (red) increases more rapidly than the size of the inner nuclei (blue). Proportional cytoplasmic volumes (defined as cytoplasmic volumes separated by the midpoints of two adjacent nuclear centers or the cell end) of the multinucleated cell are overlaid in color (bottom panel). (B) Scatter plot of nuclear size and proportional cytoplasmic volumes were measured at the end of interphase (blue: inner nuclei, red: outer nuclei). (C) cdc11-119 klp2Δ cells were grown at 36.5°C for 4.5 h. Then nuclei were displaced by centrifugation causing uneven distribution. Images taken 2 h after nuclear displacement show that nuclei which are surrounded by a larger proportional cytoplasmic volume are bigger. (D) An additional growth zone as formed in the cdc11-119 tea1Δ klp2Δ mutant increases nuclear size in its proximity. Left, overlay of DIC and cut11-GFP signal. Nuclei are numbered. (E) Average nuclear volume from four cells is plotted for every numbered nucleus. Error bars, SD. Bar, 5 μm. (F) cdc11-119 cells were arrested at 36.5°C for 9 h (4 cell cycles). Arrowheads on fluorescence image (inverted lookup table) point to representative large and small nuclei within the same cell. Bar, 10 μm.
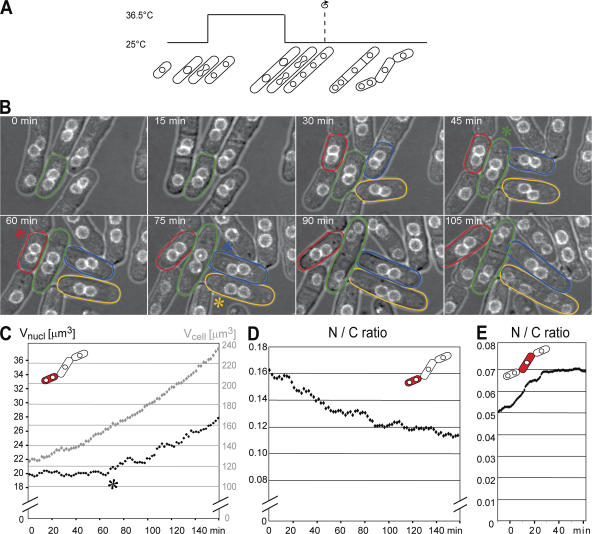
We next artificially perturbed N/C ratio to investigate how rapidly nuclear growth responds to changes in the N/C ratio. Centrifugation of multinucleated cells followed by septation generates cells with increased and decreased N/C ratios (Daga and Chang, 2005; Fig. 5 A; Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1). In cells that had a twofold elevated N/C ratio, nuclear growth was arrested and only resumed once a threshold of ∼1.5-fold the normal N/C ratio had been attained (Fig. 5, B–D). In contrast, in cells that had a reduced N/C ratio, rapid nuclear growth restored an almost normal N/C ratio, usually within less than 1 h (Fig. 5 E). Together, these experiments establish that nuclear and cell growth are not directly coupled and that nuclear size is causally dependent upon cell size. These perturbation experiments further demonstrated that during vegetative interphase growth nuclei can grow faster than the cell, but are unable to contract, even at high N/C ratios.
Figure 5.
The cell can sense and correct changed N/C ratios. (A) N/C ratios are manipulated by block and release of cytokinesis using the cdc11-119 klp2Δ mutant. Before septa form, nuclei are displaced by centrifugation resulting in cells with reduced and elevated N/C ratios. (B) Time points from Video 2. A Gaussian blur (r = 2 pixels) was applied to the fluorescence channel. Kinetics of N/C ratio correction were monitored by 3D time-lapse microscopy (2-min intervals). Individual cells with a decreased N/C ratio are marked with colored lines; an asterisk marks the approximate frame when nuclear growth starts. (C and D) Quantitation of cell size and nuclear size and corresponding N/C ratio for a high N/C ratio. Time is indicated starting at cytokinesis. The graphs represent the median of 10-min moving averages from three independent cells. (E) Low N/C ratios get corrected more rapidly than high ratios. Video 2 is available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1.
Nuclear growth in general can be either driven by an increase in nuclear volume or by an increase in NE surface area, or by a combination of both. Whereas growth of the NE requires the availability and the targeting of newly formed membrane components from the ER, growth driven by volume increase would involve either nucleocytoplasmic transport or diffusion of smaller molecules through nuclear pores and sequestering within the nucleus. Two sets of experiments suggest that the NE expansion is a result rather than the cause of nuclear volume increase. First, it has been shown that NE-ER over-proliferation is not sufficient to increase nuclear size, but instead leads to an accumulation of NE sheets around the nucleus (Lum and Wright, 1995; Tange et al., 2002). Second, when blocking nuclear export of a subset of proteins for 90–150 min using leptomycin B (LMB), a specific inhibitor of the exportin crm1, nuclear size and the N/C ratio increase by 50% (Matsuyama et al., 2006; Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1). This suggests that nucleocytoplasmic transport directly or indirectly alters nuclear size control, and contrasts with data from budding yeast, where 5–30 min of treatment with LMB had shown no obvious effect on nuclear size (Jorgensen et al., 2007). The differences in the results may be due to the more extended time course of drug treatment in our experiments. We further tested if the distribution of nuclear pores influences the N/C ratio. Cells deleted for nup133b and marked with the nucleoporin Nup107-GFP have less evenly distributed nuclear pore complexes (Bai et al., 2004), but the N/C ratio is not affected (unpublished data).
It is possible that nuclear volume could be controlled by some surrogate, such as amount of RNA or protein, numbers of ribosomes, or membrane content. In motoneurons and hepatocytes, cell size and nuclear size both correlate with the cellular RNA/DNA ratio, the expression of ribosomal genes, and general transcription rate (Sato et al., 1994; Schmidt and Schibler, 1995). Future studies will be required to dissect the molecular basis of nuclear size control in fission yeast.
A similar general cellular control that regulates nuclear growth in response to the amount of cytoplasm surrounding the nucleus may influence nuclear growth in other eukaryotes. However, differences in the cellular differentiation state and organismal developmental stage or the presence of a nuclear lamina, add more layers to N/C ratio control. Although we have shown that DNA content does not directly influence nuclear size, it might set a minimum to the size of the nucleus as suggested by the nucleoskeletal theory (Cavalier-Smith, 1982; Gregory, 2005), especially in small cells such as spores. For example, whereas wild-type spores have an N/C ratio of 0.076 ± 0.016 (see Fig. 1, A, B, and D), wee1ts spores have a 20% smaller cell size but only 8% smaller nuclei, indicating that a minimal nuclear size may have been reached (wee1-50/wee1-50, n = 136, N/C = 0.089 ± 0.017).
Nuclear size regulation could be influenced by several cellular functions such as nucleocytoplasmic transport, lipid metabolism, or ribosome biogenesis. It is important that the biochemical mechanisms underlying NE growth take into account the global cellular control we have described here, which so precisely relates growth in nuclear volume to growth of the cell. Because membrane-bound organelles are an essential part of the function and architecture of the eukaryotic cell, understanding how nuclear growth is regulated is likely to be informative about how the growth of other membranous structures within the cell are coordinated with changes in cell growth and differentiation.
Materials and methods
Strains
S. pombe strains are described in Table I. Strains were generated by genetic crosses and tested by segregation of markers or PCR.
Table I. Fission yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| PN3779 | h+ cut11-GPF∷ura4+ ura4-D18 leu1-32 | Nurse lab collection |
| PN10345 | H− cut11-GPF∷ura4+ ura4-D18 | This study |
| PN10342 | h−/h− cut11-GPF∷ura4+/cut11-GPF∷ura4+ ura4-D18/ura4-D18 leu1-32/leu1-32 | This study |
| PN10268 | h+ wee1-50 cut11-GPF∷ura4+ ura4-D18 leu1-32 | This study |
| PN10269 | h− wee1-50 cut11-GPF∷ura4+ ura4-D18 leu1-32 | This study |
| PN3781 | h+ cdc10-V50 cut11-GPF∷ura4+ ura4-D18 | Nurse lab collection |
| PN10271 | h− cdc25-22 cut11-GPF∷ura4+ ura4-D18 leu1-32 | This study |
| PN10417 | h+ cdc13Δ∷ura4+ pREP45-cdc13+(sup3-5) cut11-GPF∷ura4+ ura4-D18 ade6-704 leu1-32 | This study |
| PN10421 | orb2-34 cut11-GPF∷ura4+ ura4-D18 | This study |
| PN10422 | orb6-25 cut11-GPF∷ura4+ ura4-D18 | This study |
| PN10424 | h+ cdc11-119 klp2Δ∷ura4+ tea1Δ∷ura4+ cut11-GPF∷ura4+ ura4-D18 | This study |
| PN10419 | h+ clp1-GFP-NLS∷ura4+ cut11-GPF∷ura4+ ura4-D18 his7-336 | This study |
| PN10420 | h+ clp1-GFP-NLS∷ura4+ cdc25-22 cut11-GPF∷ura4+ ura4-D18 his7-336 | This study |
| SPV33 | h+ nup107-GFP∷KanMX ade6-M216 ura4-D18 leu1-32 | Bai et al., 2004 |
| SPV72 | h? nup107-GFP∷KanMX nup133bΔ∷KanMX ade6- ura4-D18 leu1-32 | Bai et al., 2004 |
Growth and media
Standard media and methods were used (Moreno et al., 1991; Hayles and Nurse, 1992). Unless indicated differently, all yeast strains were grown in YE4S medium at 25°C. Temperature-sensitive strains were shifted from 25 to 36.5°C for the time indicated. For nitrogen starvation, exponentially growing cells were washed and resuspended in EMM-N and grown for 12 h before microscopy. For sporulation, strains were pregrown in YE4S. The mixed cells were plated on SPA plates and incubated for 2 d at 25°C before microscopy.
Microscopy
S. pombe strains were grown in YE4S to OD595 0.2–0.4 and mounted on agarose pads (1.4% agarose in YE4S). For time-lapse microscopy, coverslips were sealed with VALAP (Vaseline, Lanolin, Paraffin; 1:1:1). Population based microscopy was performed at 23–25°C on a microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a CoolsnapHQ camera (Roper Scientific). Data were acquired using the 100× PlanFluar NA 1.45 objective taking 12 z-sections with 0.3-μm spacing. Time-lapse microscopy was performed on a microscope (Axiovert 200; Carl Zeiss, Inc.) with a spinning-disk confocal head (UltraView; Perkin-Elmer), a cooled CCD camera (Orca ER; Hamamatsu), and the 63× PlanApo NA 1.4 objective at 32°C (wild type) or 36°C (cdc11-119).
Image analysis and quantification
Images were acquired in MetaMorph (MDS Analytical Technologies) and analyzed in ImageJ (W. Rasband; National Institutes of Health, Bethesda, MD). Projections of the fluorescence channel were combined with DIC images. Cells were measured by hand assuming simple geometries, and volumes were calculated based on axial symmetries (cell: rod; nucleus: prolate ellipsoid). Statistical analyses were performed in Excel and KaleidaGraph. Box-and-Whisker plots represent the distribution, the boxes delimiting the median, the first and third quartiles, and whiskers marking 5th and 95th percentiles. The Pearson product moment correlation coefficient (r) was used to describe linear correlations.
Cdc13 switch-off
Protocol was modified from Hayles et al. (1994). PN10417 was grown on EMM-4S plates for 2 d and inoculated into EMM-N at 2 × 106 cells/ml and starved for 12 h at 25°C. Cells were then collected by centrifugation and resuspended in YE4S+Thiamine at a concentration of 106 cells/ml and were grown at 32°C. FACS samples were collected every 2 h, and aliquots mounted on agarose pads for live microscopy or fixed for quantitative DAPI staining and morphometric analysis.
Quantitative DAPI staining
15 ml of cells (10-h time point) were fixed for 20 min at 32°C with 3% formaldehyde, washed 2× with PEM and resuspended in PEMS. Cells were permeabilized by 1-h digestion with Zymolyase (0.5 mg/ml 100T in PEMS) washed for 10 min with PEMS containing 0.5% Triton X-100 at 4°C. Cells were washed with PEM and PBS and stained with 4,6-diamidino- 2-phenylindole in PBS. For microscopy cells were washed with PBS, air-dried on a glass slide, and mounted in 90% glycerol containing 1 μg/ml phenylenediamine.
Nuclear displacement
The method was adapted from Daga and Chang (2005). Nuclei of cdc11-119 arrested cells were displaced by 4-min centrifugation at 16,000 g in a microcentrifuge (5415 D; Eppendorf).
Online supplemental material
Figure S1 shows the relation between volumes of the nucleolus, nucleus, and cell for growing cells (wild-type and cdc25-22). Figure S2 shows that treatment with LMB for 90 or 150 min increases proportional nuclear size in wild-type and cdc25-22 cells. Video 1: time-lapse analysis of growing fission yeast illustrates that the N/C ratio is constant throughout the cell cycle. Time points of a selected field are shown in Fig. 3 A. Video 2 shows how the cell responds to artificial changes of the N/C ratio for small cells containing two nuclei. Selected time points are shown in Fig. 5 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708054/DC1.
Supplemental Material
Acknowledgments
We thank A. North (Rockefeller University Bio-Imaging Resource Center) for assistance with spinning disk confocal microscopy; D. McCollum and V. Doye for strains; M. Rout and D. Riveline for helpful discussions; and the members of the Nurse lab for critical comments on the manuscript.
F.R. Neumann was supported by a fellowship from the Swiss National Science Foundation. The Nurse lab is supported by grants from the Breast Cancer Research Foundation and the Rockefeller University.
Abbreviations used in this paper: DIC, differential interference contrast; LMB, leptomycin B; N/C, nuclear/cell; NE, nuclear envelope; ts, temperature sensitive.
References
- Bai, S.W., J. Rouquette, M. Umeda, W. Faigle, D. Loew, S. Sazer, and V. Doye. 2004. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 24:6379–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri, T. 1905. Zellenstudien V. Über die Abhängigkeit der Kerngrösse und Zellenzahl bei Seeigellarven von der Chromosomenzahl der Ausganszellen. Jena. Z. Naturw. 39:445–524. [Google Scholar]
- Brandt, A., F. Papagiannouli, N. Wagner, M. Wilsch-Brauninger, M. Braun, E.E. Furlong, S. Loserth, C. Wenzl, F. Pilot, N. Vogt, et al. 2006. Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr. Biol. 16:543–552. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 1982. Skeletal DNA and the evolution of genome size. Annu. Rev. Biophys. Bioeng. 11:273–302. [DOI] [PubMed] [Google Scholar]
- Daga, R.R., and F. Chang. 2005. Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA. 102:8228–8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, S.M. 2002. Visualizing chromatin dynamics in interphase nuclei. Science. 296:1412–1416. [DOI] [PubMed] [Google Scholar]
- Gerassimow, J. 1902. Die Abhängigkeit der Zelle von der Menge ihrer Kernmasse. Zeitschr. Allg. Physiol. 1:220–258. [Google Scholar]
- Gregory, T. 2005. Genome size evolution in animals. In The Evolution of the Genome. Vol. 1. T. Gregory, editor. Elsevier Academic Press, London. 4–87.
- Hayles, J., and P. Nurse. 1992. Genetics of the fission yeast Schizosaccharomyces pombe. Annu. Rev. Genet. 26:373–402. [DOI] [PubMed] [Google Scholar]
- Hayles, J., D. Fisher, A. Woollard, and P. Nurse. 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2- mitotic B cyclin complex. Cell. 78:813–822. [DOI] [PubMed] [Google Scholar]
- Hertwig, R. 1903. Ueber die Korrelation von Zell-und Kerngrösse und ihre Bedeutung für die Geschlechtliche Differenzierung und die Teilung der Zelle. Biologisches Centralblatt. 23:4–62. [Google Scholar]
- Höög, J.L., C. Schwartz, A.T. Noon, E.T. O'Toole, D.N. Mastronarde, J.R. McIntosh, and C. Antony. 2007. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell. 12:349–361. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., N.P. Edgington, B.L. Schneider, I. Rupes, M. Tyers, and B. Futcher. 2007. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 8:3523–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovtchev, G., V. Schubert, A. Meister, M. Barow, and I. Schubert. 2006. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114:77–82. [DOI] [PubMed] [Google Scholar]
- Lum, P.Y., and R. Wright. 1995. Degradation of HMG-CoA reductase-induced membranes in the fission yeast, Schizosaccharomyces pombe. J. Cell Biol. 131:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F. 2004. Cellular length control systems. Annu. Rev. Cell Dev. Biol. 20:677–693. [DOI] [PubMed] [Google Scholar]
- Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, et al. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24:841–847. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823. [DOI] [PubMed] [Google Scholar]
- Nurse, P. 1975. Genetic control of cell size at cell division in yeast. Nature. 256:547–551. [DOI] [PubMed] [Google Scholar]
- Nurse, P. 1985. The genetic control of cell volume. In The Evolution of Genome Size. T. Cavalier-Smith, editor. John Wiley & Sons, Ltd., Chichester, UK. 185–196.
- Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. [DOI] [PubMed] [Google Scholar]
- Orgel, L.E., and F.H. Crick. 1980. Selfish DNA: the ultimate parasite. Nature. 284:604–607. [DOI] [PubMed] [Google Scholar]
- Russell, P., and P. Nurse. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 45:145–153. [DOI] [PubMed] [Google Scholar]
- Sato, S., S.B. Burgess, and D.L. McIlwain. 1994. Transcription and motoneuron size. J. Neurochem. 63:1609–1615. [DOI] [PubMed] [Google Scholar]
- Schmidt, E.E., and U. Schibler. 1995. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J. Cell Biol. 128:467–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, V., and P. Nurse. 1994. Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 13:2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., F. Hediger, F.R. Neumann, and S.M. Gasser. 2004. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38:305–345. [DOI] [PubMed] [Google Scholar]
- Tange, Y., A. Hirata, and O. Niwa. 2002. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 115:4375–4385. [DOI] [PubMed] [Google Scholar]
- Umen, J.G. 2005. The elusive sizer. Curr. Opin. Cell Biol. 17:435–441. [DOI] [PubMed] [Google Scholar]
- Verde, F., J. Mata, and P. Nurse. 1995. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R.R., E.V. Vaisberg, R. Ding, P. Nurse, and J.R. McIntosh. 1998. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 9:2839–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., D. Yarar, T.H. Giddings Jr., and D.N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg, J., and M.M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7:9–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.