Abstract
DNA replication in eukaryotic cells is tightly controlled by a licensing mechanism, ensuring that each origin fires once and only once per cell cycle. We demonstrate that the ataxia telangiectasia and Rad3 related (ATR)–mediated S phase checkpoint acts as a surveillance mechanism to prevent rereplication. Thus, disruption of licensing control will not induce significant rereplication in mammalian cells when the ATR checkpoint is intact. We also demonstrate that single-stranded DNA (ssDNA) is the initial signal that activates the checkpoint when licensing control is compromised in mammalian cells. We demonstrate that uncontrolled DNA unwinding by minichromosome maintenance proteins upon Cdt1 overexpression is an important mechanism that leads to ssDNA accumulation and checkpoint activation. Furthermore, we show that replication protein A 2 and retinoblastoma protein are both downstream targets for ATR that are important for the inhibition of DNA rereplication. We reveal the molecular mechanisms by which the ATR-mediated S phase checkpoint pathway prevents DNA rereplication and thus significantly improve our understanding of how rereplication is prevented in mammalian cells.
Introduction
To maintain genome stability, DNA replication occurs only once in each cell cycle. This is achieved by the cell cycle–dependent licensing control that permits assembly of prereplication complexes (pre-RCs) at replication origins only once per cell cycle (Mendez and Stillman, 2003; Diffley, 2004; Nishitani and Lygerou, 2004; Blow and Dutta, 2005; Arias and Walter, 2007). On exit from metaphase, the minichromosome maintenance (MCM) 2–7 complex is recruited to replication origins to establish pre-RCs. Once replication is activated by the Cdks and Cdc7/Dbf4 kinase, pre-RCs are disassembled and the reassociation of MCM proteins with origins is not permitted until the completion of mitosis.
Regulation of Cdt1 activity during the cell cycle is critical for the prevention of reassembly of pre-RCs within a single cell cycle (Blow and Dutta, 2005; Arias and Walter, 2007). When cells enter S phase, Cdt1 is ubiquitinated and degraded (Nishitani et al., 2001; Li et al., 2003; Zhong et al., 2003; Arias and Walter, 2005), and this cell cycle–regulated Cdt1 degradation is important for the prevention of rereplication. In the Xenopus laevis cell free system, the addition of Cdt1 in G2 extracts induces rereplication, which is enhanced in the absence of geminin (Arias and Walter, 2005; Li and Blow, 2005; Maiorano et al., 2005; Yoshida et al., 2005). In mammalian cells, overexpression of Cdt1 promotes substantial rereplication in certain tumor cell lines (Vaziri et al., 2003; Melixetian et al., 2004; Zhu et al., 2004).
Rereplication of the genome, or even a segment of it, could initiate gene amplification and cause chromosomal translocation and loss, contributing to the tumorigenesis process. It has been shown that cell cycle checkpoints are activated when rereplication is induced (Machida and Dutta, 2005). In yeast, rereplication leads to accumulation of extensive double-stranded breaks (DSBs) and activates checkpoints (Archambault et al., 2005; Green and Li, 2005). In the X. laevis cell free system, a caffeine-sensitive phosphorylation of Chk1 occurs when rereplication is induced (Li and Blow, 2005). In mammalian cells, overexpression of Cdt1 or the loss of geminin activates the G2/M checkpoint or induces apoptosis (Vaziri et al., 2003; Melixetian et al., 2004; Zhu and Dutta, 2006). Recently, by using X. laevis egg extracts, it was demonstrated that extensive rereplication, but not a single round of rereplication, leads to head-to-tail collision of rereplicating forks resulting in DNA fragmentation and checkpoint activation (Davidson et al., 2006). This finding suggests an important mechanism underlying checkpoint activation when uncontrolled rereplication is induced. However, other mechanisms that do not require extensive rereplication likely exist. In mammalian cells, when licensing control is disrupted by Cdt1 overexpression, intriguingly, extensive rereplication only occurs in certain tumor cell lines (Vaziri et al., 2003; Tatsumi et al., 2006; this paper). In primary cell lines and several tumor cell lines, significant rereplication is not induced but checkpoints are activated.
Two protein kinases, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR), play critical roles in sensing abnormal DNA structures and initiating signal transduction in the checkpoint pathways (Abraham, 2001; Shiloh, 2001). Although related, ATM and ATR respond to DNA damage in overlapping but distinct pathways. ATM responds mainly to DSBs, whereas ATR is activated by various kinds of DNA damage, including UV and hydroxyurea treatment, which stall replication forks. ATR activation requires DNA replication, which is different from ATM activation (Michael et al., 2000; Lupardus et al., 2002; Tercero et al., 2003).
In this study, we demonstrated that in mammalian cells, the ATR-mediated S phase checkpoint is activated at the onset of rereplication before the appearance of DSBs. This ATR-mediated S phase checkpoint acts directly to inhibit rereplication so that significant rereplication is prevented in the cell lines with an intact ATR pathway. We also demonstrated that uncontrolled DNA unwinding initiated by MCM proteins from relicensed origins leads to single-stranded DNA (ssDNA) accumulation, revealing an important mechanism underlying checkpoint activation in the absence of extensive rereplication. Unlike ATR, ATM does not play critical roles in the prevention of rereplication but coordinates with ATR to activate the G2/M checkpoint. Finally, we identified important effector proteins, including RPA2 and retinoblastoma protein (Rb), downstream of ATR to mediate the inhibition of rereplication. We propose that specific defects in the ATR-mediated S phase checkpoint pathways present in certain tumor cell lines contribute to their susceptibility to rereplication.
Results
ATR plays a critical role in the prevention of rereplication
In mammalian cells, when licensing control is disrupted by Cdt1/Cdc6 overexpression, it is intriguing that substantial rereplication occurs only in certain tumor cell lines but not in primary cell lines and several other tumor cell lines (Vaziri et al., 2003; Tatsumi et al., 2006). Although p53 is involved (Vaziri et al., 2003), the loss of geminin-induced rereplication occurs in a p53-independent manner (Melixetian et al., 2004; Zhu et al., 2004), suggesting that pathways other than p53 are also important for preventing rereplication.
Consistent with previous findings (Vaziri et al., 2003; Tatsumi et al., 2006), we observed that overexpression of Cdt1 by adenovirus infection was sufficient to induce DNA rereplication in H1299, HeLa, and U2OS tumor cells, but failed to cause overt rereplication in many other cell lines, including primary cell lines IMR90 and WI38 and tumor cell lines T98G and A549 (Fig. 1 A). By using a higher titer of Ad-Cdt1 viruses, Cdt1 overexpression levels in these rereplication-susceptible cell lines were similar to those in U2OS and H1299 cell lines (Fig. 1 A and not depicted). Co-overexpression of Cdc6 with Cdt1 also failed to induce overt rereplication in T98G, A549, and IMR90 cells, although it enhanced Cdt1-induced rereplication in U2OS and H1299 cells (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200704138/DC1; and not depicted).
Figure 1.
Cdt1 overexpression leads to checkpoint activation in the absence of detectable rereplication. (A) H1299, U2OS, T98G, and IMR90 cells were infected by adenoviruses encoding GFP (Ad-Vec) or human Cdt1 (Ad-Cdt1) with the indicated viral titers: H1299 (107 pfu/ml, MOI = 5; or 5 × 107 pfu/ml, MOI = 25), U2OS (3 × 107 pfu/ml, MOI = 15; or 108 pfu/ml, MOI = 50), T98G (6 × 108 pfu/ml, MOI = 460), and IMR90 (6 × 108 pfu/ml, MOI = 600). The cell cycle profile was analyzed by FACS analysis 48 h after adenovirus infection. The percentage of cells containing more than 4 N of DNA content is indicated in each panel. The Cdt1 expression levels are shown by Western blot analysis using the same amount of cell lysates. (B) Cell extracts were prepared from H1299 (5 × 107 pfu/ml, MOI = 25), A549 (6 × 108 pfu/ml, MOI = 400), U2OS (5 × 107 pfu/ml, MOI = 25), and IMR90 (6 × 108 pfu/ml, MOI = 600) cells (left) or from IMR90 cells expressing shRNA specific for ATR or ATM (middle and right) 48 h after adenoviral infection with Ad-Vec or Ad-Cdt1. Western blot analyses were performed using antibodies as indicated. p-Chk1, phosphorylation at S317; p-Chk2, phosphorylation at T68.
Although extensive rereplication was not observed in T98G, A549, and IMR90 cells, checkpoints were activated after Cdt1 overexpression. Chk1 and 2 were both phosphorylated upon Ad-Cdt1 infection in an ATR- and ATM-dependent manner, respectively (Fig. 1 B), although the phosphorylation levels in IMR90 and A549 were reduced compared with H1299 and U2OS cells, where rereplication was achieved extensively (Fig. 1 B, left; and not depicted). These data suggest that both the ATM–Chk2 and ATR–Chk1 pathways are activated when Cdt1 is overexpressed, even in the absence of extensive rereplication, although overt rereplication may amplify damage signals and enhance checkpoint activation.
The critical role of cell cycle checkpoint activation is to maintain genome stability. The observation that ATR and ATM were activated by Cdt1 overexpression in the cell lines without extensive rereplication prompted us to examine whether checkpoint activation in fact prevented rereplication. Because Cdt1 overexpression–induced ATR and ATM activation was observed in rereplication-susceptible cell lines such as U2OS and H1299 (Fig. 1 B), we anticipated that defects allowing rereplication in these cell lines were present at the effector pathways downstream of ATR and/or ATM. We first examined rereplication levels in U2OS cells when the expression of ATR or ATM was suppressed by short hairpin RNA (shRNA). If the susceptibility of U2OS cells to Cdt1-induced rereplication was indeed caused by a defect in one branch of the ATR and/or ATM pathways, inactivation of ATR and/or ATM would influence multiple pathways, thereby leading to more severe rereplication after Cdt1 overexpression. We used a low titer of Ad-Cdt1 viruses, which did not induce obvious rereplication in U2OS cells. Interestingly, when ATR-shRNA was expressed, robust rereplication was induced after Ad-Cdt1 infection, whereas inhibition of ATM did not lead to more rereplication (Fig. 2 A). The Cdt1 expression levels were similar in all the cell lines that were examined (Fig. 2 A, bottom). Consistently, inhibition of endogenous ATR kinase activity by overexpressing kinase-dead (KD) Flag-ATR in U2OS cells (Nghiem et al., 2002) also led to higher levels of DNA rereplication after Ad-Cdt1 infection (Fig. 2 B). As an alternative approach, we inactivated DDB1 and Cdt2, the components of a Cul4 ubiquitin ligase that are required for Cdt1 ubiquitination and degradation (Hu et al., 2004; Jin et al., 2006) by shRNA in U2OS cells. Silencing the expression of DDB1 or Cdt2 leads to detectable accumulation of Cdt1 (1.5- to 2-fold more) and rereplication (Fig. 2 C). Inactivation of ATR by shRNAs before the depletion of DDB1 or Cdt2 significantly enhanced the rereplication levels compared with vector-infected, ATR-proficient cells. Collectively, these data suggest that ATR, but not ATM, plays an important role in the suppression of DNA rereplication.
Figure 2.
Inactivation of ATR but not ATM leads to enhanced rereplication after Cdt1 overexpression. (A) U2OS cells expressing ATM-shRNA, ATR-shRNA, or vector MKO were infected with Ad-vec (GFP) or Ad-Cdt1 (2 × 107 pfu/ml, MOI = 10), and FACS analysis was performed 48 h after infection (top). Cdt1 overexpression was shown by Western blot analysis (bottom). Actin was used as a loading control. (B) U2OS cell lines carrying tet-on Flag-ATR wild type or Flag-ATR-KD were treated with 1 μg/ml doxycyclin 24 h before Ad-vec or Ad-Cdt1 (5 × 107 pfu/ml, MOI = 25) infection. FACS analysis was performed 48 h after adenoviral infection. The expression of Cdt1 was shown by Western blot analysis. (C) U2OS cells were infected with retroviruses encoding ATR-shRNA or a vector, followed by inactivation of DDB1 or Cdt2 by shRNA. A week later, after drug selection to enrich shRNA-DDB1– or shRNA-Cdt2–expressing cells, FACS analysis was performed (left). The expression of Cdt1 and DDB1 was examined by Western blot analysis (right). (D) ATR or ATM was silenced by expressing shRNAs in A549, T98G, and IMR90 cells. FACS analysis was performed 48 h after indicated adenoviral infection. Western blot analyses were performed as indicated. (E) The expression of Chk1 and 2 was inhibited by expressing shRNA in U2OS and A549 cells and revealed by Western blot analysis. FACS analysis was performed 48 h after Ad-vec or Ad-Cdt1 infection. The adenovirus titers U2OS (2 × 107 pfu/ml, MOI = 10) and A549 (6 × 108 pfu/ml, MOI = 400) were used. Black lines indicate that intervening lanes have been spliced out.
We then tested whether ATR is required to prevent rereplication in the cell lines that are resistant to Cdt1-induced rereplication. Strikingly, when ATR activity was inhibited in IMR90, WI38, A549, and T98G cell lines by shRNAs, a significant amount of cells underwent rereplication after Cdt1 or Cdt1/Cdc6 was overexpressed (Fig. 2 D and not depicted). Therefore, ATR acts as a critical barrier in these cell lines to resist Cdt1-induced rereplication. In contrast, inhibition of ATM activity did not disrupt the cellular control that prevents rereplication (Fig. 2 D). These results uncover a critical function of ATR in the prevention of rereplication, especially when licensing control is impaired.
Chk1 and 2 are two effector protein kinases downstream of ATR and ATM, respectively. We suppressed the expression of Chk1 and 2 in multiple cell lines and examined the levels of rereplication after Cdt1 overexpression. Inactivation of Chk1, but not Chk2, significantly increased the rereplication levels in U2OS cells and also led to rereplication in A549 cells, which were normally resistant to Cdt1-induced rereplication (Fig. 2 E). These data support the finding that the ATR–Chk1 pathway, not the ATM–Chk2 pathway, is involved in the suppression of rereplication.
Activation of the ATR–Chk1 pathway occurs before that of the ATM–Chk2 pathway when Cdt1 is overexpressed
During DNA rereplication, both DSBs and ssDNA are accumulated (Melixetian et al., 2004; Archambault et al., 2005; Green and Li, 2005; Davidson et al., 2006; Zhu and Dutta, 2006). We observed that DNA lesions were also accumulated in the cell lines where Cdt1 overexpression failed to induce significant rereplication. In every cell line tested, RPA1 and 2 were loaded onto chromatin when Cdt1 was overexpressed, suggesting that ssDNA accumulated on chromatin (Fig. 3 A). RPA2 was also phosphorylated on chromatin, revealed by the presence of slower migrating forms of RPA2 (Fig. 3 A). The presence of ssDNA was confirmed by BrdU immunostaining under nondenaturing conditions (Fig. 3 B). Likewise, we also detected H2AX phosphorylation, a common marker for indicating DSBs (Burma et al., 2001), in the cell lines without obvious rereplication, including IMR90, T98G, and A549 cells (Fig. 3 C and not depicted). These data suggest that Cdt1 overexpression leads to the generation of both ssDNA and DSBs even in the absence of extensive rereplication.
Figure 3.
Cdt1 overexpression–induced rereplication leads to the accumulation of ssDNA and DSBs. (A) H1299 (5 × 107 pfu/ml, MOI = 25), A549 (6 × 108 pfu/ml, MOI = 400), U2OS (5 × 107 pfu/ml, MOI = 25), T98G (6 × 108 pfu/ml, MOI = 460), and IMR90 (6 × 108 pfu/ml, MOI = 600) cells were infected with Ad-vec or Ad-Cdt1. Chromatin and cell lysates were prepared 48 h after infection. Western blot analyses were performed as indicated. (B) IMR90 cells were infected with Ad-vec or Ad-Cdt1 (6 × 108 pfu/ml, MOI = 600) for 48 h and cells were labeled with BrdU during the last 24 h of infection. Cells were fixed under nondenaturing condition and stained with anti-BrdU antibodies. DAPI was used for nuclear staining. Cells treated with 0.5 μg/ml DNase 1 before fixing were used as a positive control. Similar experiments were performed using U2OS cells. The percentage of BrdU-positive cells after Ad-Cdt1 infection in IMR90 and U2OS cells is indicated. Bar, 20 μm. (C) Cell extracts were prepared from H1299, A549, U2OS, and IMR90 cells 48 h after Ad-vec or Ad-Cdt1 infection as described in A. Western blot analysis was performed using antibody recognizing γ-H2AX. Actin was used as a loading control. (D) Chromatin and cell lysates were prepared at different time points after U2OS cells were infected with Ad-vec or Ad-Cdt1 (5 × 107 pfu/ml, MOI = 25). Western blot analyses were performed as indicated. Phospho-Chk1, phosphorylated at S317; phospho-Chk2, phosphorylated at T68; un-RPA2, unphosphorylated RPA2; p-RPA2, phosphorylated RPA2. Cell cycle profiles were shown at different time points after Ad-vec or Ad-Cdt1 infection. Black lines indicate that intervening lanes have been spliced out.
To investigate how DNA lesions were generated when Cdt1 was overexpressed, we performed a time course analysis to monitor the accumulation of ssDNA and DSBs. When a lower titer of Ad-Cdt1 was used, a slower kinetics of Cdt1 overexpression and Chk1 activation was observed than when a higher titer of Ad-Cdt1 was used (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200704138/DC1). Therefore, to clearly demonstrate the temporal order of ssDNA and DSB accumulation and checkpoint activation, we used the low titer of Ad-Cdt1 or Ad-vec (5 × 107 pfu/ml, MOI = 25) to infect U2OS cells. At the 12-h time point (4 h after significant Cdt1 expression), although no obvious rereplication was detected (Fig. 3 D, bottom), Chk1 was already phosphorylated at the ATR phosphorylation site S317 (Fig. 3 D, left). At an approximately similar time, RPA chromatin loading was observed (Fig. 3 D, right), suggesting that ssDNA was generated. Interestingly, Chk2 phosphorylation at ATM phosphorylation site T68 was not detected until the 24-h time point, when rereplication was initially detected. At the same time point, H2AX was phosphorylated, suggesting that DSBs were likely generated relatively late. When ATR was inactivated in U2OS cells by shRNA, the kinetics of Chk1, Chk2, and H2AX phosphorylation was altered. Chk1 phosphorylation was delayed because of the critical role of ATR to phosphorylate Chk1, whereas ATM-mediated Chk2 and H2AX phosphorylation as well as RPA chromatin loading occurred earlier (Fig. S3). This is consistent with the idea that rereplication is more profound in ATR-deficient cells, which leads to more severe DNA lesions and checkpoint activation. These data also suggest that the temporal order of the accumulation of phosphorylated species of Chk1 and 2 and H2AX observed in ATR-proficient cells (Fig. 3 D) is not likely caused by different sensitivities of the specific antibodies.
Collectively, these data suggest that ATR is activated before ATM at an early stage when Cdt1 is overexpressed, and ssDNA, which accumulates before DSBs, likely serves as an early signal to activate ATR. The activation of ATR at an early stage after Cdt1 overexpression but before significant rereplication is induced is consistent with the observation that ATR plays an important role in preventing rereplication.
The S phase checkpoint mediates ATR activation and prevents DNA rereplication
The observation of ssDNA accumulation at early stages after Cdt1 overexpression suggests that ATR is likely activated by the same S phase checkpoint pathway as when replication forks are stalled by DNA-damaging agents. Indeed, we observed that Rad17 was phosphorylated on chromatin, and Rad9 chromatin loading was promoted when Cdt1 was overexpressed in multiple cell lines, including the cell lines without significant rereplication (Fig. 4 A). In addition, Cdt1 overexpression– induced Chk1 phosphorylation was diminished when the expression of ATR-interacting protein (ATRIP) or Rad17 was inhibited by shRNA (not depicted). Meanwhile, inhibition of Rad17 or ATRIP expression caused more severe rereplication in U2OS cells and led to rereplication in A549 cells, which were otherwise resistant to Cdt1-induced rereplication (Fig. 4 B). These data demonstrate a critical role of the ATR-mediated S phase checkpoint pathway in both sensing the loss of licensing control and preventing rereplication, which highlights a new aspect of the S phase checkpoint in the maintenance of genome stability.
Figure 4.
Rad17 and ATRIP are required for preventing Cdt1 overexpression–induced rereplication. (A) Chromatin was purified from indicated cell lines 48 h after Ad-vec or Ad-Cdt1 infection (T98G: 6 × 108 pfu/ml, MOI = 460; IMR90: 6 × 108 pfu/ml, MOI = 600; U2OS: 5 × 107 pfu/ml, MOI = 25; A549: 6 × 108 pfu/ml, MOI = 400). Western blot analyses were performed using antibodies recognizing Rad9, Rad17, and phosphorylated Rad17 at S645. (B) Rad17 and ATRIP were silenced by shRNA in U2OS and A549 cells. The protein levels of Rad17 or ATRIP were shown by Western blot analysis. FACS analysis was performed using indicated cell lines 48 h after Ad-vec or Ad-Cdt1 infection.
DNA unwinding by MCM proteins from relicensed origins contributes to the accumulation of ssDNA
It is important to understand how ssDNA is accumulated at early stages after Cdt1 overexpression to activate ATR. To understand which steps of rereplication are involved in checkpoint activation, we sought to overexpress Cdt1 after S phase so that we could inhibit different steps of rereplication without interfering with normal S phase replication.
U2OS cells were synchronized by a double thymidine block. 6 h after releasing, nocodazole was added and after another 6 h, almost all cells exited from S phase, as revealed by the cell cycle profile and low BrdU incorporation (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200704138/DC1; and not depicted). At this time point, cells were infected with Ad-Cdt1 or Ad-vec (Fig. 5 A). 24 h after adenoviral infection, both attached and round-up cells were collected after manually shaking off. Phosphorylation of Cdk1 at Tyr15 and cyclin A expression were detected in attached cells, and the mitotic marker phosphohistone H3 was present in shaken-off cells (Fig. S4 B), suggesting that a significant number of attached cells are in G2 phase, which is consistent with previous findings (Ballabeni et al., 2004). Cdt1 overexpression was predominantly observed in attached cells (Fig. S4 B), indicating that detached mitotic cells either were difficult to infect or expressed Cdt1 at low levels. Attached cells were harvested for FACS analysis after Ad-Cdt1 or Ad-vec infection and a significant amount of rereplication was induced after Ad-Cdt1 infection (Fig. 5 A). This Cdt1- induced rereplication was prevented when adenoviruses encoding geminin, p27, or a Cdc7 KD mutant (Cdc7KD and Cdc7-D196N; Tsuji et al., 2006) were used to infect cells before Ad-Cdt1 infection (Fig. 5 B), suggesting that Cdt1-mediated licensing and the kinase activity of Cdk and Cdc7 are required for rereplication induced after S phase. Cdt1-induced rereplication was also inhibited when cells were treated with aphidicolin, an inhibitor of DNA polymerase α (Fig. 5 B; Sheaff et al., 1991).
Figure 5.
ssDNA accumulation is likely caused by aberrant DNA unwinding mediated by MCM proteins after Cdt1 overexpression. (A) U2OS cells were synchronized by double thymidine block and 25 ng/ml nocodozole was added into the medium 6 h after releasing. After another 6 h, cells were infected with Ad-vec or Ad-Cdt1 (3 × 108 pfu/ml, MOI = 75) or nothing. After shaking-off of mitotic cells, attached cells were collected 24 h after infection for FACS and Western blot analyses using phospho-Chk1 (S317)– and phospho-Chk2 (T68)–specific antibodies. (B) U2OS cells were synchronized as described in A. 12 h after releasing cells from thymidine block with the presence of 25 ng/ml nocodozole for the last 6 h, cells were infected with Ad-vec (vec), Ad-geminin (gem), Ad-p27 (p27), or Ad-Cdc7KD (Cdc7KD), or treated with 1 μg/ml aphidicolin (Aphi). 3 h after the first adenoviral infection or aphidicolin treatment, cells were reinfected with Ad-vec or Ad-Cdt1 (3 × 108 pfu/ml, MOI = 75). After mitotic shaking-off, attached cells were collected 24 h after the first adenoviral infection. Cell cycle profile was monitored by FACS analysis. (C) Chromatin and whole cell lysates were prepared from attached cells at the same time points as when FACS was performed as described in B. Western blot analyses were performed using antibodies as indicated. (D) Chromatin was purified as described in C and Western blot analysis was performed using antibodies recognizing MCM2. MCM2p, phosphorylated MCM2. (E) Synchronized U2OS cells as described in B were infected with Ad-vec or Ad-MCM7-K387A (3 × 108 pfu/ml, MOI = 75) 3 h before Ad-vec or Ad-Cdt1 infection (3 × 108 pfu/ml, MOI = 75). Chromatin and whole cell extracts were purified 24 h after first adenoviral infection. Western blot analyses were performed as indicated. Cell cycle profile was monitored by FACS analysis.
Similar to asynchronized cells, checkpoint was activated when Cdt1 was overexpressed after S phase (Fig. 5 A, bottom). Overexpression of geminin, p27, and Cdc7KD before Cdt1 prevented ssDNA accumulation and attenuated checkpoint activation (Fig. 5 C). Interestingly, after aphidicolin treatment, although rereplication was prevented to similar levels as when p27 or Cdc7KD was overexpressed (Fig. 5 B), ssDNA accumulation shown by RPA1 chromatin loading was still evidently accompanied by checkpoint activation (Fig. 5 C), although the level of checkpoint activation was reduced. These data suggest that the initiation of DNA rereplication, which requires Cdks and Cdc7 activity, is important for generating ssDNA and activating checkpoint. Because aphidicolin inhibits DNA polymerase α and thus likely arrests replication/rereplication forks close to replication origins, continuous DNA synthesis and extensive rereplication seem to not necessarily be required for ssDNA accumulation and checkpoint activation, although they may possibly enhance these events.
When Cdt1/Cdc6 is overexpressed, MCM proteins are reloaded onto chromatin, but reloaded MCM proteins may not coordinate properly with other replication proteins. For instance, MCM-mediated DNA unwinding may exceed the availability of polymerases that can be used to synthesize DNA when origins are refired. This may cause functional uncoupling of MCM helicase and DNA polymerases leading to the accumulation of ssDNA, which has been described previously when DNA polymerase α is inactivated or the replication fork is stalled (Walter and Newport, 2000; Byun et al., 2005). If this model is correct, the MCM helicase activity that is required for DNA unwinding at the onset of rereplication would be needed for ssDNA accumulation and checkpoint activation upon Cdt1 overexpression.
When p27 or Cdc7KD was overexpressed before Cdt1/Cdc6 overexpression, we noticed that the loss of ssDNA accumulation and checkpoint activation was accompanied by a decrease of MCM2 phosphorylation, as demonstrated by a decrease of the faster migrating band (the phosphorylated MCM2 species) on SDS-PAGE gel (Fig. 5 D). Because this cell cycle–regulated MCM2 phosphorylation is believed to be important for the helicase activities of the MCM complex (Tsuji et al., 2006), this finding supports the idea that MCM-mediated DNA unwinding is involved in generating ssDNA.
To more directly examine whether DNA unwinding at relicensed origins was required for generating ssDNA, we inhibited MCM helicase activity by overexpressing an MCM7 mutant (K387A) carrying a mutation at its conserved ATPase motif that is required for replication helicase activity (You et al., 1999; You et al., 2002). Immunoprecipitation showed that MCM7-K387A interacted with other endogenous MCM proteins (not depicted) and that overexpression of this mutant prevented cells from entering S phase, suggesting a dominant-negative effect of this mutant (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200704138/DC1). As shown in Fig. 5 E, overexpression of the MCM-K387A mutant before Cdt1 overexpression in synchronized cells that had completed S phase inhibited rereplication and significantly reduced RPA chromatin loading and Chk1 phosphorylation. These data strongly suggest that MCM-mediated DNA unwinding from relicensed origins is required for generating ssDNA and activating the checkpoint when Cdt1 is overexpressed.
ATR phosphorylates RPA2 and MCM2 when Cdt1 is overexpressed
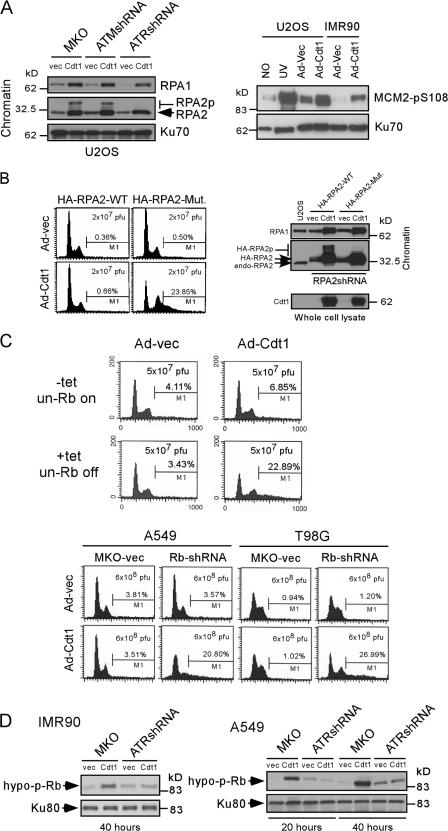
We have demonstrated that the ATR-mediated checkpoint can be activated by the initial steps of rereplication, which acts to prevent further rereplication. What is the mechanism by which activated ATR inhibits rereplication? We observed that RPA2, which plays essential roles in replication initiation and elongation (Binz et al., 2004; Fanning et al., 2006), was phosphorylated when Cdt1 was overexpressed (Fig. 3 A) in an ATR- but not ATM-dependent manner (Fig. 6 A, left). Similarly, phosphorylation of MCM2 at S108, a site phosphorylated by ATR upon UV and hydroxyurea treatment (Cortez et al., 2004), was also observed after Cdt1 overexpression (Fig. 6 A, right).
Figure 6.
RPA2, MCM2, and Rb are important effector proteins downstream of ATR to inhibit Cdt1-induced rereplication. (A) 48 h after Ad-vec or Ad-Cdt1 infection, chromatin was purified from U2OS cells expressing shRNA-ATR, shRNA-ATM, or vector MKO (left), or cell lysates were prepared from U2OS or IMR90 cells (right). The adenovirus titers U2OS (5 × 107 pfu/ml, MOI = 25) and IMR90 (6 × 108 pfu/ml, MOI = 600) were used. After 1 h, 50 J/m2 UV treatment was used as a positive control for MCM2 phosphorylation. Western blot analyses were performed as indicated. RPA2p, phosphorylated RPA2; MCM2-pS108, phosphorylated MCM2 at S108. (B) U2OS cell lines expressing the HA-RPA2 wild type or HA-RPA2-phospho mutant (S4A/S8A/S11A/S12A/S13A/T21A/S33A) with endogenous RPA2 silenced by shRNA were generated as described previously (Olson et al., 2006). FACS analyses were performed 48 h after Ad-vec or Ad-Cdt1 infection (5 × 107 pfu/ml, MOI = 25). Western blot analyses were performed as indicated. endo-RPA2, endogenous RPA2; HA-RPA2p, phosphorylated HA-tagged RPA2. (C) U2OS cells carrying tetracycline-regulated constitutively active Rb were infected with Ad-vec or Ad-Cdt1 (5 × 107 pfu/ml, MOI = 25) in the presence of tetracycline (+tet, un-Rb off) or 24 h after removal of tetracycline (−tet, un-Rb on). 36 h after infection, cell cycle profiles were monitored by FACS analysis (top). Rb was silenced in A549 and T98G cells by expressing shRNA from retroviral vector MKO. FACS analysis was performed 48 h after Ad-vec or Ad-Cdt1 infection (bottom; A549: 6 × 108 pfu/ml, MOI = 400; T98G: 6 × 108 pfu/ml, MOI = 460). (D) Cell lysates were prepared from IMR90 or A549 cells expressing ATR-shRNA or vector MKO 20 or 40 h after Ad-vec or Ad-Cdt1 infection (6 × 108 pfu/ml; IMR90, MOI = 600; A549, MOI = 400). Rb phosphorylation was analyzed by the antibody G99-549, which specifically recognizes nonphosphorylated S608 species. Ku80 was used as a loading control.
Multiple damage-inducible and ATR-dependent phosphorylation sites have been identified at the N terminus of RPA2 (Zernik-Kobak et al., 1997; Nuss et al., 2005; Olson et al., 2006). Cdt1 overexpression–induced RPA2 phosphorylation was completely abolished in the HA-tagged RPA2 mutant carrying S/T-to-A substitutions at the ATR-dependent phosphorylation sites S4/S8/S11-13/T21/S33 (Fig. 6 B, right; Olson et al., 2006). Meanwhile, rereplication was also significantly enhanced in the cell line expressing the HA- RPA2-phospho mutant with endogenous RPA2 silenced by shRNA compared with the cell line expressing the HA- RPA2 wild type (Fig. 6 B, left). These results suggest that ATR-mediated RPA2 phosphorylation plays a direct role in suppressing DNA rereplication. Because MCM2 S108 is not the only ATR site and not all ATR phosphorylation sites on MCM2 have been identified (Cortez et al., 2004), a direct role of MCM2 phosphorylation by ATR in the suppression of rereplication is currently difficult to analyze.
Rb plays an important role in the prevention of rereplication
Overexpression of Cdt1 or Cdt1/Cdc6 can readily induce rereplication in H1299 and U2OS cells. Because checkpoint activation is intact in these two cell lines, the defects in suppressing rereplication are probably present at downstream steps. As described previously, the susceptibility of H1299 cells to rereplication is caused by a loss of p53 function (Vaziri et al., 2003). However, the U2OS cell line carries wild-type p53, and its genetic basis for permitting rereplication is not clear.
A defect in Rb dephosphorylation was observed in U2OS cells during progression from mitosis into G1 (Broceno et al., 2002). Upon DNA damage, Rb dephosphorylation is induced (Linke et al., 1997; Martelli and Livingston, 1999; Lan et al., 2002; Avni et al., 2003). To examine whether a dephosphorylation defect of Rb in U2OS cells might contribute to Cdt1-induced rereplication, we used a U2OS cell line that expressed constitutively active Rb under the control of tetracycline (Chew et al., 1998; Broceno et al., 2002). In this Rb mutant, 14 of the 15 conserved Cdk consensus sites are mutated. Cdt1 overexpression–induced rereplication was significantly inhibited when the expression of hypophosphorylated Rb was induced (Fig. 6 C, top). These data suggest that a defect in U2OS cells affecting Rb dephosphorylation is likely responsible for permitting Cdt1-induced rereplication. Consistent with an important role of Rb in the suppression of rereplication, when the expression of Rb was inhibited by shRNA in T98G and A549 cells, substantial rereplication was observed after Cdt1 overexpression (Fig. 6 C, bottom).
To assess whether overexpression of Cdt1 leads to Rb dephosphorylation and thus inhibition of rereplication, we monitored Rb dephosphorylation by using the antibody G99-549, which specifically recognizes the hypophosphorylated state of Rb after Cdt1 overexpression (Broceno et al., 2002; Avni et al., 2003). In both IMR90 and A549 cell lines, Rb dephosphorylation was induced when Cdt1 was overexpressed (Fig. 6 D). Interestingly, when ATR expression was inhibited by shRNA, accumulation of hypophosphorylated Rb species was diminished after Cdt1 overexpression. Collectively, these data suggest that ATR-mediated checkpoint activation induces Rb dephosphorylation, which is involved in the inhibition of rereplication induced by Cdt1 overexpression.
Both ATM and ATR contribute to the activation of the G2/M checkpoint induced by Cdt1 overexpression
Although both the ATM and ATR pathways are activated when Cdt1 is overexpressed, only the ATR pathway is necessary for the suppression of rereplication. What is the biological role of ATM activation? Previous papers have shown that Cdt1 overexpression activates the G2/M checkpoint by arresting cells before mitosis and that ATR contributes to this arrest (Vaziri et al., 2003; Zhu and Dutta, 2006). As illustrated in Fig. 7, after Cdt1 overexpression, cells expressing ATM-shRNA were also partially impaired in G2/M arrest as were ATR-shRNA–expressing cells, whereas simultaneous inhibition of both ATM and ATR further increased the number of cells that escaped G2/M arrest. Thus, ATR activation at an early stage after Cdt1 overexpression directly inhibits DNA rereplication and ATM is subsequently activated, which acts synergistically with ATR to arrest cells in G2/M.
Figure 7.
Down-regulation of ATR and/or ATM impairs G2/M checkpoint in response to Cdt1 overexpression. Immunostaining using anti– phosphohistone H3 (mitotic maker) antibodies was performed 48 h after Ad-vec or Ad-Cdt1 infection (5 × 107 pfu/ml, MOI = 25) in U2OS cells expressing vector MKO, shRNA-ATR, shRNA-ATM, or both shRNA-ATR and shRNA-ATM. In each experiment, >300 cells were counted. SD was derived from three independent experiments.
Discussion
In this study, we demonstrated that the ATR-mediated S phase checkpoint provides a protection mechanism beyond licensing control to prevent rereplication and maintain genome stability. We also demonstrated that in mammalian cells, one initial signal to trigger checkpoint activation by Cdt1 overexpression is ssDNA accumulation, which is caused by uncontrolled MCM-mediated DNA unwinding, whereas DSBs are generated subsequently. This finding suggests a novel mechanism to activate the checkpoint at the initiation of rereplication before uncontrolled rereplication occurs.
MCM-mediated DNA unwinding from relicensed origin is important for the activation of the checkpoint
The head-to-tail collision model illustrates the mechanism by which DSBs are generated during overt rereplication (Davidson et al., 2006). This model suggests that replication forks may chase one another when uncontrolled rereplication occurs, leading to fork collision and DNA fragmentation. However, in mammalian cells, the checkpoint can be activated in the absence of extensive DNA rereplication, suggesting that initiation of a limited amount of rereplication in mammalian cells is sufficient to activate the checkpoint. We also observed that in the presence of aphidicolin, the checkpoint is activated, although rereplication is effectively inhibited. Because rereplication forks are arrested in the presence of aphidicolin, replication and rereplication forks should not actively encounter each other and the initial signals that activate the checkpoint in mammalian cells are not likely generated by head-to-tail fork collision as in the X. laevis cell free system (Davidson et al., 2006). Another mechanism involving certain steps at the onset of rereplication is likely used to initiate checkpoint activation in mammalian cells.
Suppression of Cdk or Cdc7 activity abrogates ssDNA accumulation and checkpoint activation when rereplication is induced after S phase, suggesting that the reassembly of pre-RC per se is not sufficient for checkpoint activation and that the initiation of DNA rereplication is required. Further studies demonstrated that the inhibition of MCM helicase activities before Cdt1 overexpression prevents ssDNA accumulation and checkpoint activation, which supports the notion that DNA unwinding by MCM proteins from relicensed origins is involved in generating ssDNA and activating the checkpoint.
DNA replication is a highly coordinated process, and reinitiation of DNA replication within the same cell cycle disrupts this balance. When MCM proteins are reloaded onto chromatin, MCM-mediated DNA unwinding may exceed the rate or capacity of DNA polymerases to synthesize DNA, leading to ssDNA accumulation. It is known that primases synthesize RNA primers at a relative slow rate (Sheaff and Kuchta, 1993; Swart and Griep, 1995). DNA polymerases for lagging strand synthesis are limited (Langston and O'Donnell, 2006). Replication proteins, such as MCM10 and DNA polymerase α/primase (Nasheuer et al., 1991; Izumi et al., 2001), are often phosphorylated during the cell cycle, and such modifications may serve to suppress their replication activities at an inappropriate time. The uncoupling of MCM helicase and polymerase activities was proposed as an important mechanism leading to the generation of ssDNA when replication forks are stalled by DNA-damaging agents (Walter and Newport, 2000; Byun et al., 2005). Our data suggest that the initiation of rereplication uses similar uncoupling mechanisms to activate checkpoint control, thereby providing a molecular explanation of how the conventional ATR-mediated S phase checkpoint is activated upon loss of licensing control.
Based on our findings, we propose that Cdt1 overexpression leads to reassembly of pre-RCs and that unscheduled DNA unwinding from relicensed origins by MCM proteins is uncoupled from DNA synthesis, leading to ssDNA accumulation and ATR activation (Fig. 8). DSBs generated subsequently are probably caused by the collapse of stalled rereplication forks.
Figure 8.
Possible mechanisms leading to the generation of DNA lesions when Cdt1 is overexpressed.
Although in mammalian cells head-to-tail fork collision may not be the primary cause for checkpoint activation, uncontrolled rereplication induced by Cdt1 in the absence of an intact checkpoint control, as seen in U2OS or H1299 cells, could lead to fork collision and generate more DNA lesions. Therefore, the DNA unwinding model and the head-to-tail fork collision model, respectively, address the mechanisms underlying checkpoint activation at the initial stage of rereplication and when rereplication occurs for multiple rounds. During uncontrolled rereplication, in addition to fork collision as described previously (Davidson et al., 2006), DSBs may also be generated when new rereplication forks encounter Okazaki fragments at the existing forks (Fig. 8).
The ATR-mediated checkpoint acts to prevent rereplication when licensing control is impaired
Although both ATR and ATM pathways are activated by Cdt1 overexpression, our studies suggest that the ATR but not the ATM pathway is essential to prevent rereplication when the licensing control is compromised. ATR is activated earlier than ATM, before rereplication is detected. This quick response implies that ATR detects abnormal chromosomal structures generated at the initial steps of rereplication and suppresses rereplication immediately after it is activated. The inhibition of rereplicaion can be achieved by suppressing DNA rereplication initiation from other origins or elongation at existing rereplication forks. ATM, however, is activated at a much later stage, presumably as a consequence of processing of the initial lesions that generate DSBs.
Inhibition of DNA rereplication by the ATR-mediated S phase checkpoint is essential for the prevention of genome instability. During a normal cell cycle, mistakes may occur to allow reassembly of pre-RCs at one or more origins. Inhibition of rereplication immediately after DNA unwinding would prevent rereplication forks from running into existing forks to generate DNA fragments, which can be highly recombinogenic and cause chromosomal rearrangement. This inhibition would also limit rereplication to a minimal extent, possibly right after the synthesis of RNA/DNA primers or small stretches of DNA at origins, so that checkpoint-activated repair pathways would be capable of effectively removing these duplicated sequences. It has been described that Rad51 is recruited to repair rereplication-associated lesions (Zhu and Dutta, 2006). The G2/M checkpoint activated by ATR ensures that DNA lesions are repaired and a normal licensing control is reestablished before cells reenter the cell cycle. In contrast, when the loss of licensing control persists or is chronic, as under our experimental conditions or because of somatic mutations, rereplication forks are stalled by the continuous reloading of MCM proteins. Secondary DNA lesions, such as DSBs, are generated by fork collapse, activating ATM. Under such conditions, ATM activation is important for repairing DSBs and maintaining G2/M arrest until DNA lesions are repaired.
Multiple downstream effector pathways are activated to suppress rereplication
We demonstrate that rereplication is suppressed by the ATR-mediated S phase checkpoint pathway and that multiple ATR downstream effector proteins appear to be involved in mediating this suppression. Thus, susceptibility to rereplication can be caused by various mutations in multiple pathways downstream of ATR, which provides a molecular basis for the difference in rereplication among different mammalian cell lines when licensing control is disrupted.
Both RPA2 and MCM2, two proteins essential for replication, were phosphorylated at ATR-dependent phosphorylation sites after Cdt1 overexpression. Mutating RPA2 N-terminal damage-inducible phosphorylation sites led to significant rereplication when the licensing control was impaired. Damage-induced RPA2 phosphorylation at these sites is proposed to prevent RPA from migrating into replication centers in S phase, thus attenuating the replication function of the RPA complex (Vassin et al., 2004; Olson et al., 2006). The phosphorylation of MCM proteins at the ATR consensus sites (Cortez et al., 2004) may also directly inhibit MCM functions required for DNA replication.
Besides targeting the replication machinery directly, ATR also inhibits rereplication indirectly through regulating requisite proteins that link checkpoints with cell cycle progression. p53 was proposed to inhibit rereplication through the p53–p21–Cdk pathway upon checkpoint activation (Vaziri et al., 2003). In this study, we identified Rb as another downstream effector protein in the ATR pathway and showed that a defect in Rb function is the cause for the susceptibility of U2OS cells to Cdt1 overexpression–induced rereplication.
Hypophosphorylated Rb binds to the E2F/DP family proteins and sequesters their transcription activities (Harbour and Dean, 2000). Cdks, cyclins, and several replication proteins such as DNA polymerase α, ORC1, MCM proteins, Cdc6, and Cdt1 are E2F-regulated genes (Ohtani et al., 1996; Herwig and Strauss, 1997; Helin, 1998; Leone et al., 1998; Yan et al., 1998; Yoshida and Inoue, 2004). In addition, Rb directly interacts with replication proteins such as MCM7, DNA polymerase α, and replication factor C (Takemura et al., 1997; Sterner et al., 1998; Pennaneach et al., 2001; Gladden and Diehl, 2003). Recent studies also demonstrated that DNA damage induces Rb dephosphorylation, and hypophosphorylated Rb binds to replication origin proximal sites specifically after DNA damage (Linke et al., 1997; Martelli and Livingston, 1999; Lan et al., 2002; Avni et al., 2003). Thus, hypophosphorylated Rb likely inhibits DNA replication through multiple mechanisms (Knudsen et al., 1998; Angus et al., 2004). Although Rb dephosphorylation was described after DNA damage, we further clarified the mechanism of this checkpoint response by showing that Rb dephosphorylation is under the control of ATR. We postulate that the accumulation of hypophosphorylated Rb species is caused by Cdk down-regulation when the ATR-mediated S phase checkpoint is activated.
DNA rereplication can be a major driving force for genome amplification and genetic instability, which are highly associated with cancer development. We suggest that multiple effector pathways downstream of ATR, including Rb, are used to effectively suppress rereplication in mammalian cells when licensing control is impaired. Both p53 and Rb are tightly linked to cancer prevention. Therefore, our studies highlight a new aspect of S phase checkpoint control in the maintenance of genome stability and suppression of tumorigenesis through its critical role in the prevention of rereplication.
Materials and methods
Cell culture and retroviral infection
U2OS, H1299, A549, T98G, and IMR90 cells were grown in DME supplemented with 10% fetal bovine serum. To induce the expression of Flag-ATR wild type or Flag-ATR-KD in the established U2OS cell lines (Nghiem et al., 2002), 1 μg/ml doxycycline was added to the media for 24 h. U2OS cells with tetracycline-regulated constitutive active Rb (provided by S. Mittnacht, Institute of Cancer Research, London, UK; Chew et al., 1998) were maintained in medium, supplemented with 2 μg/ml tetracycline, 300 μg/ml G418, and 0.5 μg/ml puromycin. Rb expression was induced by culturing cells in tetracycline-free medium for 24 h.
Stable U2OS cell lines expressing an HA-tagged RPA2 or phospho-RPA2 mutant were generated by retroviral infection using pBabe vector as described previously (Olson et al., 2006). Silencing of endogenous RPA2 in these cells was conducted by two rounds of retroviral infection using a pMKO vector (Masutomi et al., 2003) that expresses the RPA2 shRNA target sequence located in the 3′ untranslated region of the mRNA, followed by drug selection (Vassin et al., 2004). Other retroviral infection was also performed accordingly.
Whole cell lysate and chromatin isolation
Cells were lysed in NETN (150 mM NaCl, 1 mM EDTA, 20 mM Tris-Cl, pH 8.0, and 0.5% NP-40) containing protease and phosphatase inhibitors (Sigma-Aldrich). For chromatin isolation, cells were washed with PBS, resuspended in CSK buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgClB2, 1 mM EGTA, 50 mM Na-F, 0.1 mM Na-orthovanadate [Sigma-Aldrich], 0.1% Triton X-100 [Sigma-Aldrich], and protease inhibitors), and incubated on ice for 10 min. Cytoplasmic proteins were separated from nuclei by low speed centrifugation at 1,300 g for 5 min. Isolated nuclei were washed once in CSK buffer and lysed in solution (3 mM EDTA, 0.2 mM EGTA, 1mM DTT, and protease inhibitors). After centrifugation (1,700 g for 5 min), pellets were resuspended in CSK buffer. 2× SDS loading buffer was added and samples were boiled for 10 min.
Plasmids
shRNA retroviral plasmids were constructed by inserting annealed shRNA oligos into a pMKO retroviral vector (Masutomi et al., 2003). The shRNA sequences used were ATM (GCACCAGTCCAGTATTGGCTT and AACATACTACTCAAAGACATT), ATR (CGAGACTTCTGCGGATTGCAG and AACCTCCGTGATGTTGCTTGA; Casper et al., 2002), ATRIP (AAGGTCCACAGATTATTAGAT; Ball et al., 2005), Chk2 (CAGTGTCCACTCAGGAACTCT), Chk1 (AAGCGTGCCGTAGACTGTCCA and AAGTACTCCAGTTCTCAGCCA), claspin (CCTTGCTTAGAGCTGAGTCTT and GGAAAGAAAGGCAGCCAGATT; Chini and Chen, 2003), Rad17 (CAGACTGGGTTGACCCATCTT; Zou et al., 2002), DDB1 (TGAGTGCTTGACATACCTTGA), and Cdt2 (GTTCCTGGTGAACTTAAACTT; Jin et al., 2006). Sequence information for Chk1 and 2 shRNA was provided by A. Maclaren, J. Scorah, and C. McGowan (The Scripps Research Institute, La Jolla, CA).
To obtain the MCM7-K387A mutant, site-directed mutagenesis was conducted by QuickChange mutagenesis (Stratagene) using full-length MCM7 as a template. The oligonucleotides 5′-CTGATGGGGGATCCTGGTGTGGCCGCGTCTCAGCTCCTGTCATACATTGAT-3′ and 5′-ATCAATGTATGACAGGAGCTGAGACGCGGCCACACCAGGATCCCCC-ATCAG-3′ were used as primers.
The Cdc7KD was generated by mutating D196 to N in the conserved kinase motif of Cdc7 (Tsuji et al., 2006).
Adenovirus construction and infection
Adenoviruses encoding GFP, human Cdt1, Cdc7KD, Cdc6, and MCM7-K387A were generated by using the AdEasy system (He et al., 1998). In brief, the target cDNAs were constructed into a pAd-track-CMV shuttle vector and in vivo recombination was performed by transforming the pAd-track-CMV shuttle vector together with the pAd-Easy-1 adenoviral vector into a BJ5813 competent cell by electroporation. The recombinant adenoviral plasmids were transfected into 293 cells to produce corresponding recombinant adenoviruses. Large-scale purification of viruses was obtained by CsCl density gradient centrifugation. Recombinant adenoviruses encoding p27 and geminin were provided by S. Reed (The Scripps Research Institute, La Jolla, CA) and J. Cook (University of North Carolina at Chapel Hill, Chapel Hill, NC).
To enrich cells that have completed S phase, U2OS cells were synchronized by double thymidine block (2 mM thymidine for 16 h and no thymidine for 12 h). 6 h after being release from the second thymidine block, 25 ng/ml nocodozole was added to the media, and after another 6 h, cells were infected with adenoviruses. After mitotic shaking-off, attached cells were collected for further analysis.
Infection of T98G cells in G0 was performed by culturing cells in a medium containing 0.1% fetal bovine serum for 24 h, followed by adenoviral infection of Ad-vec or Ad-MCM7-K387A. 24 h after adenoviral infection, cells were released from G0 by adding 10% fetal bovine serum into medium.
Immunofluorescence staining and antibodies
To examine G2/M arrest, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and stained with antibodies recognizing phosphohistone-H3 (the mitotic marker), followed by DNA staining with DAPI. For measuring ssDNA formation, cells were labeled with 10 μM BrdU for 24 h after adenoviral infection. Cells were fixed, permeabilized, and stained with anti-BrdU antibodies.
The following antibodies were used in this study: Cdt1 was produced as described previously (Li et al., 2003); the phospho-Chk1 (S317) antibody was obtained from R&D Systems; phospho-Chk2 (T68) was obtained from Cell Signaling Technology; Chk1, Chk2, Ku70, Rad17, Rad9, and geminin antibodies were obtained from Santa Cruz Biotechnology, Inc.; p27, actin, RPA1, RPA2, ATM, and ATR antibodies were obtained from EMD; γ-H2AX and phosphohistone H3 antibodies were obtained from Millipore; ATRIP and phospho-MCM2 (S108) antibodies were obtained from Bethyl Laboratories, Inc.; Rb, cyclinA, cyclinE, MCM2, and DDB1 antibodies were obtained from BD Biosciences. Cdc7 antibody was provided by T. Tsuji and W. Jiang (The Burnham Institute for Medical Research, La Jolla, CA).
FACS analysis
Cells were collected and fixed with ice cold 70% ethanol overnight at 4°C. After washing with PBS, cells were stained with 15 μg/ml propidium iodide solution containing 38 mM sodium citrate and 10 μg/ml RNase A and analyzed on a flow cytometer (Becton Dickinson) using Cellquest software (BD Biosciences). For BrdU incorporation analysis, cells were pulse labeled with 10 mM BrdU for 1 h. After fixation with ice cold 70% ethanol for 1 h, cells were resuspended in 2 mM HCl and incubated for 20 min at room temperature. After centrifugation, cell pellets were resuspended in 0.5 ml of 0.1 M Na2B4O7, pH 8.5, and after a PBS wash, cells were stained with 50 μl of antibody solution (30 μl PBS containing 0.5% Tween-20/0.5% BSA + 1 μl mouse anti-BrdU monoclonal antibody) for 1 h at room temperature, followed by 30-min staining with FITC-conjugated secondary antibody in the dark before propidium iodide staining.
Online supplemental material
Fig. S1 shows that overexpression of Cdt1 or Cdt1/Cdc6 induces DNA rereplication in certain tumor cell lines. Fig. S2 shows that Cdt1 overexpression and Chk1 phosphorylation were detected earlier when a higher titer of Ad-Cdt1 was used. Fig. S3 shows a time course analysis of checkpoint activation and RPA chromatin loading in ATR-deficient or -proficient cells after Cdt1 overexpression. Fig. S4 shows that U2OS cells were synchronized to G2/M. Fig. S5 shows that overexpresion of the MCM7-K387A mutant prevents S phase entry. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704138/DC1.
Acknowledgments
We thank Ann Maclaren, Jenny Scorah, and Clare McGowan for sharing with us the Chk1 and 2 RNAi sequences. We thank Toshiya Tsuji and Wei Jiang for helping us generate the Cdc7KD mutant and providing us the Cdc7 antibody. We thank Steven Reed for the p27 adenoviruses, Jeanette Cook for the geminin adenoviruses, and William Hahn for the MKO vector. We thank Sibylle Mittnacht for the U2OS cell lines expressing tetracycline-regulated wild-type Rb or constitutively active Rb. We thank Steven Reed, Ann Maclaren, and Christian Nievera for critical reading of this manuscript.
This work was supported by National Institutes of Health grant CA102361 and an Ellison Medical Foundation New Scholar award (AG-NS-0251-04) to X. Wu.
E. Liu and A.Y-L. Lee contributed equally to this paper.
E. Liu's present address is Signal Transduction Program, Burnham Institute for Medical Research, La Jolla, CA 92037.
T. Chiba's present address is Dept. of Investigative Pathology, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki 852-8523, Japan.
Abbreviations used in this paper: ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; ATRIP, ATR-interacting protein; DSB, double-stranded break; KD, kinase dead; MCM, minichromosome maintenance; pre-RC, prereplication complex; Rb, retinoblastoma protein; RPA, replication protein A; shRNA, short hairpin RNA; ssDNA, single-stranded DNA.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177–2196. [DOI] [PubMed] [Google Scholar]
- Angus, S.P., C.N. Mayhew, D.A. Solomon, W.A. Braden, M.P. Markey, Y. Okuno, M.C. Cardoso, D.M. Gilbert, and E.S. Knudsen. 2004. RB reversibly inhibits DNA replication via two temporally distinct mechanisms. Mol. Cell. Biol. 24:5404–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault, V., A.E. Ikui, B.J. Drapkin, and F.R. Cross. 2005. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 25:6707–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias, E.E., and J.C. Walter. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19:114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias, E.E., and J.C. Walter. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21:497–518. [DOI] [PubMed] [Google Scholar]
- Avni, D., H. Yang, F. Martelli, F. Hofmann, W.M. ElShamy, S. Ganesan, R. Scully, and D.M. Livingston. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell. 12:735–746. [DOI] [PubMed] [Google Scholar]
- Ball, H.L., J.S. Myers, and D. Cortez. 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 16:2372–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabeni, A., M. Melixetian, R. Zamponi, L. Masiero, F. Marinoni, and K. Helin. 2004. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 23:3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz, S.K., A.M. Sheehan, and M.S. Wold. 2004. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst.). 3:1015–1024. [DOI] [PubMed] [Google Scholar]
- Blow, J.J., and A. Dutta. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broceno, C., S. Wilkie, and S. Mittnacht. 2002. RB activation defect in tumor cell lines. Proc. Natl. Acad. Sci. USA. 99:14200–14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma, S., B.P. Chen, M. Murphy, A. Kurimasa, and D.J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- Byun, T.S., M. Pacek, M.C. Yee, J.C. Walter, and K.A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, A.M., P. Nghiem, M.F. Arlt, and T.W. Glover. 2002. ATR regulates fragile site stability. Cell. 111:779–789. [DOI] [PubMed] [Google Scholar]
- Chew, Y.P., M. Ellis, S. Wilkie, and S. Mittnacht. 1998. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene. 17:2177–2186. [DOI] [PubMed] [Google Scholar]
- Chini, C.C., and J. Chen. 2003. Human claspin is required for replication checkpoint control. J. Biol. Chem. 278:30057–30062. [DOI] [PubMed] [Google Scholar]
- Cortez, D., G. Glick, and S.J. Elledge. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 101:10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, I.F., A. Li, and J.J. Blow. 2006. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell. 24:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J.F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778–R786. [DOI] [PubMed] [Google Scholar]
- Fanning, E., V. Klimovich, and A.R. Nager. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden, A.B., and J.A. Diehl. 2003. The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB.MCM7 binding. J. Biol. Chem. 278:9754–9760. [DOI] [PubMed] [Google Scholar]
- Green, B.M., and J.J. Li. 2005. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell. 16:421– 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour, J.W., and D.C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393–2409. [DOI] [PubMed] [Google Scholar]
- He, T.C., S. Zhou, L. T.da Costa, J. Yu, K.W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 95:2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin, K. 1998. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev. 8:28–35. [DOI] [PubMed] [Google Scholar]
- Herwig, S., and M. Strauss. 1997. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 246:581–601. [DOI] [PubMed] [Google Scholar]
- Hu, J., C.M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003–1009. [DOI] [PubMed] [Google Scholar]
- Izumi, M., F. Yatagai, and F. Hanaoka. 2001. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 276:48526–48531. [DOI] [PubMed] [Google Scholar]
- Jin, J., E.E. Arias, J. Chen, J.W. Harper, and J.C. Walter. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 23:709–721. [DOI] [PubMed] [Google Scholar]
- Knudsen, E.S., C. Buckmaster, T.T. Chen, J.R. Feramisco, and J.Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, Z., Z. Sever-Chroneos, M.W. Strobeck, C.H. Park, R. Baskaran, W. Edelmann, G. Leone, and E.S. Knudsen. 2002. DNA damage invokes mismatch repair-dependent cyclin D1 attenuation and retinoblastoma signaling pathways to inhibit CDK2. J. Biol. Chem. 277:8372–8381. [DOI] [PubMed] [Google Scholar]
- Langston, L.D., and M. O'Donnell. 2006. DNA replication: keep moving and don't mind the gap. Mol. Cell. 23:155–160. [DOI] [PubMed] [Google Scholar]
- Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R.S. Williams, and J.R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A., and J.J. Blow. 2005. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 24:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Q. Zhao, R. Liao, P. Sun, and X. Wu. 2003. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278:30854–30858. [DOI] [PubMed] [Google Scholar]
- Linke, S.P., M.P. Harris, S.E. Neugebauer, K.C. Clarkin, H.M. Shepard, D.C. Maneval, and G.M. Wahl. 1997. p53-mediated accumulation of hypophosphorylated pRb after the G1 restriction point fails to halt cell cycle progression. Oncogene. 15:337–345. [DOI] [PubMed] [Google Scholar]
- Lupardus, P.J., T. Byun, M.C. Yee, M. Hekmat-Nejad, and K.A. Cimprich. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 16:2327–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, Y.J., and A. Dutta. 2005. Cellular checkpoint mechanisms monitoring proper initiation of DNA replication. J. Biol. Chem. 280:6253–6256. [DOI] [PubMed] [Google Scholar]
- Maiorano, D., L. Krasinska, M. Lutzmann, and M. Mechali. 2005. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr. Biol. 15:146–153. [DOI] [PubMed] [Google Scholar]
- Martelli, F., and D.M. Livingston. 1999. Regulation of endogenous E2F1 stability by the retinoblastoma family proteins. Proc. Natl. Acad. Sci. USA. 96:2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutomi, K., E.Y. Yu, S. Khurts, I. Ben Porath, J.L. Currier, G.B. Metz, M.W. Brooks, S. Kaneko, S. Murakami, J.A. DeCaprio, et al. 2003. Telomerase maintains telomere structure in normal human cells. Cell. 114:241–253. [DOI] [PubMed] [Google Scholar]
- Melixetian, M., A. Ballabeni, L. Masiero, P. Gasparini, R. Zamponi, J. Bartek, J. Lukas, and K. Helin. 2004. Loss of geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, J., and B. Stillman. 2003. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 25:1158–1167. [DOI] [PubMed] [Google Scholar]
- Michael, W.M., R. Ott, E. Fanning, and J. Newport. 2000. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 289:2133–2137. [DOI] [PubMed] [Google Scholar]
- Nasheuer, H.P., A. Moore, A.F. Wahl, and T.S. Wang. 1991. Cell cycle-dependent phosphorylation of human DNA polymerase alpha. J. Biol. Chem. 266:7893–7903. [PubMed] [Google Scholar]
- Nghiem, P., P.K. Park, Y.S. Kim Ys, B.N. Desai, and S.L. Schreiber. 2002. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277:4428–4434. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., and Z. Lygerou. 2004. DNA replication licensing. Front. Biosci. 9:2115–2132. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., S. Taraviras, Z. Lygerou, and T. Nishimoto. 2001. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276:44905–44911. [DOI] [PubMed] [Google Scholar]
- Nuss, J.E., S.M. Patrick, G.G. Oakley, G.M. Alter, J.G. Robison, K. Dixon, and J.J. Turchi. 2005. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry. 44:8428–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani, K., J. DeGregori, G. Leone, D.R. Herendeen, T.J. Kelly, and J.R. Nevins. 1996. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 16:6977–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, E., C.J. Nievera, V. Klimovich, E. Fanning, and X. Wu. 2006. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 281:39517–39533. [DOI] [PubMed] [Google Scholar]
- Pennaneach, V., I. Salles-Passador, A. Munshi, H. Brickner, K. Regazzoni, F. Dick, N. Dyson, T.T. Chen, J.Y. Wang, R. Fotedar, and A. Fotedar. 2001. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol. Cell. 7:715–727. [DOI] [PubMed] [Google Scholar]
- Sheaff, R.J., and R.D. Kuchta. 1993. Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry. 32:3027–3037. [DOI] [PubMed] [Google Scholar]
- Sheaff, R., D. Ilsley, and R. Kuchta. 1991. Mechanism of DNA polymerase alpha inhibition by aphidicolin. Biochemistry. 30:8590–8597. [DOI] [PubMed] [Google Scholar]
- Shiloh, Y. 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11:71–77. [DOI] [PubMed] [Google Scholar]
- Sterner, J.M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J.M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart, J.R., and M.A. Griep. 1995. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 34:16097–16106. [DOI] [PubMed] [Google Scholar]
- Takemura, M., T. Kitagawa, S. Izuta, J. Wasa, A. Takai, T. Akiyama, and S. Yoshida. 1997. Phosphorylated retinoblastoma protein stimulates DNA polymerase alpha. Oncogene. 15:2483–2492. [DOI] [PubMed] [Google Scholar]
- Tatsumi, Y., N. Sugimoto, T. Yugawa, M. Narisawa-Saito, T. Kiyono, and M. Fujita. 2006. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J. Cell Sci. 119:3128–3140. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., M.P. Longhese, and J.F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 11:1323–1336. [DOI] [PubMed] [Google Scholar]
- Tsuji, T., S.B. Ficarro, and W. Jiang. 2006. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell. 17:4459–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin, V.M., M.S. Wold, and J.A. Borowiec. 2004. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 24:1930–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D.S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 11:997–1008. [DOI] [PubMed] [Google Scholar]
- Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 5:617–627. [DOI] [PubMed] [Google Scholar]
- Yan, Z., J. DeGregori, R. Shohet, G. Leone, B. Stillman, J.R. Nevins, and R.S. Williams. 1998. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA. 95:3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., and I. Inoue. 2004. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 23:3802–3812. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., H. Takisawa, and Y. Kubota. 2005. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells. 10:63–73. [DOI] [PubMed] [Google Scholar]
- You, Z., Y. Komamura, and Y. Ishimi. 1999. Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol. Cell. Biol. 19:8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Z., Y. Ishimi, H. Masai, and F. Hanaoka. 2002. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J. Biol. Chem. 277:42471–42479. [DOI] [PubMed] [Google Scholar]
- Zernik-Kobak, M., K. Vasunia, M. Connelly, C.W. Anderson, and K. Dixon. 1997. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 272:23896–23904. [DOI] [PubMed] [Google Scholar]
- Zhong, W., H. Feng, F.E. Santiago, and E.T. Kipreos. 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 423:885–889. [DOI] [PubMed] [Google Scholar]
- Zhu, W., and A. Dutta. 2006. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol. Cell. Biol. 26:4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., Y. Chen, and A. Dutta. 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24:7140–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., D. Cortez, and S.J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]