Abstract
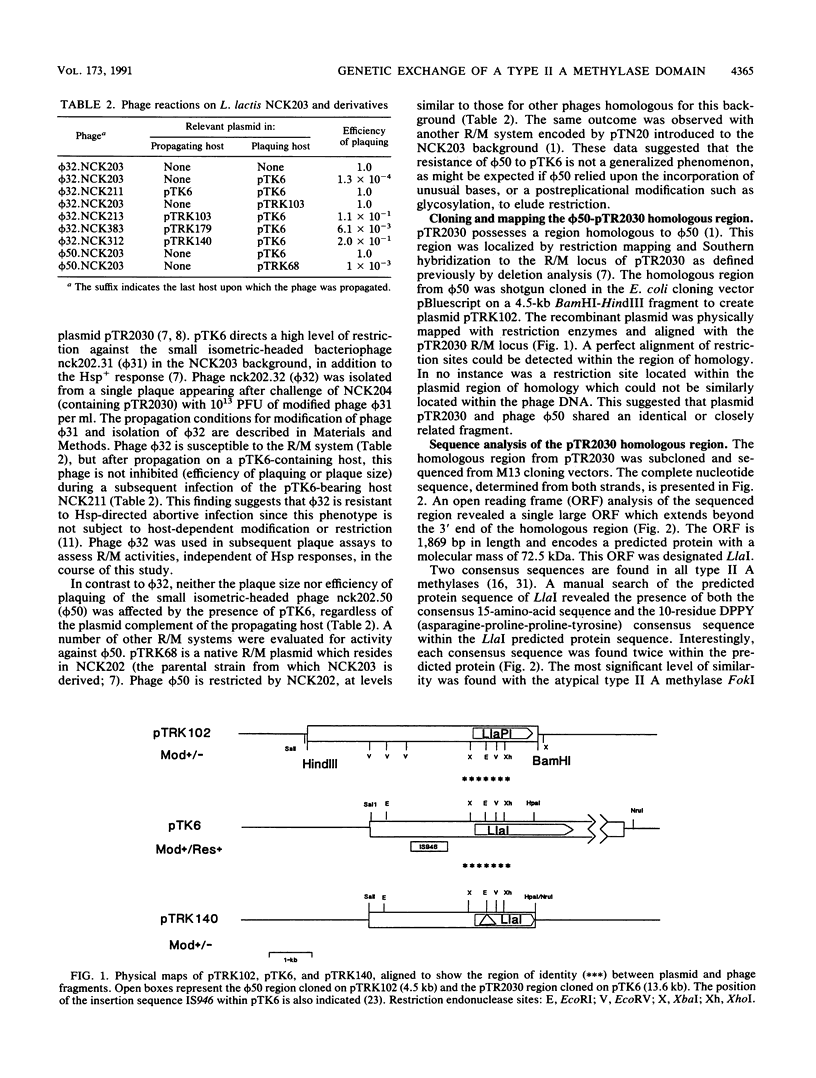
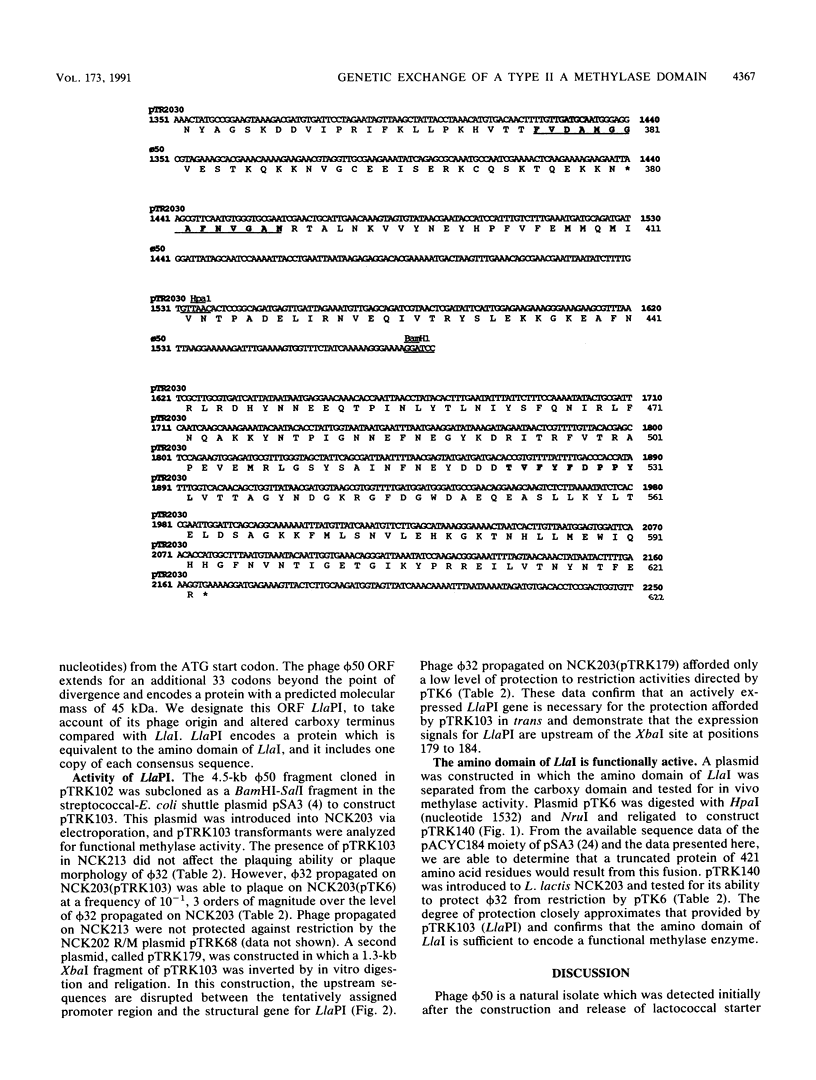
The conjugative plasmid pTR2030 confers bacteriophage resistance to lactococci by two independent mechanisms, an abortive infection mechanism (Hsp+) and a restriction and modification system (R+/M+). pTR2030 transconjugants of lactococcal strains are used in the dairy industry to prolong the usefulness of mesophilic starter cultures. One bacteriophage which has emerged against a pTR2030 transconjugant is not susceptible to either of the two defense systems encoded by the plasmid. Phage nck202.50 (phi 50) is completely resistant to restriction by pTR2030. A region of homology between pTR2030 and phi 50 was subcloned, physically mapped, and sequenced. A region of 1,273 bp was identical in both plasmid and phage, suggesting that the fragment had recently been transferred between the two genomes. Sequence analysis confirmed that the transferred region encoded greater than 55% of the amino domain of the structural gene for a type II methylase designated LlaI. The LlaI gene is 1,869 bp in length and shows organizational similarities to the type II A methylase FokI. In addition to the amino domain, upstream sequences, possibly containing the expression signals, were present on the phage genome. The phage phi 50 fragment containing the methylase amino domain, designated LlaPI, when cloned onto the shuttle vector pSA3 was capable of modifying another phage genome in trans. This is the first report of the genetic exchange between a bacterium and a phage which confers a selective advantage on the phage. Definition of the LlaI system on pTR2030 provides the first evidence that type II systems contribute to restriction and modification phenotypes during host-dependent replication of phages in lactococci.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T., Klaenhammer T. R. Molecular Characterization of Three Small Isometric-Headed Bacteriophages Which Vary in Their Sensitivity to the Lactococcal Phage Resistance Plasmid pTR2030. Appl Environ Microbiol. 1991 May;57(5):1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemaer J. P., Schrauwen P. P., Sourrouille J. L., Guy P. Multiple modification/restriction systems in lactic streptococci and their significance in defining a phage-typing system. J Dairy Res. 1980 Oct;47(3):401–409. doi: 10.1017/s0022029900021294. [DOI] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Wilkinson J., Swinton D., Schlagman S., Macdonald P. M., Mosig G. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J Bacteriol. 1985 Nov;164(2):932–937. doi: 10.1128/jb.164.2.932-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Kotani H., Sugisaki H., Takanami M. The fokI restriction-modification system. I. Organization and nucleotide sequences of the restriction and modification genes. J Biol Chem. 1989 Apr 5;264(10):5751–5756. [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Transformation of Streptococcus lactis Protoplasts by Plasmid DNA. Appl Environ Microbiol. 1982 May;43(5):1213–1215. doi: 10.1128/aem.43.5.1213-1215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landry D., Looney M. C., Feehery G. R., Slatko B. E., Jack W. E., Schildkraut I., Wilson G. G. M.FokI methylates adenine in both strands of its asymmetric recognition sequence. Gene. 1989 Apr 15;77(1):1–10. doi: 10.1016/0378-1119(89)90353-3. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Looney M. C., Moran L. S., Jack W. E., Feehery G. R., Benner J. S., Slatko B. E., Wilson G. G. Nucleotide sequence of the FokI restriction-modification system: separate strand-specificity domains in the methyltransferase. Gene. 1989 Aug 15;80(2):193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- Luchansky J. B., Muriana P. M., Klaenhammer T. R. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol Microbiol. 1988 Sep;2(5):637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Montenegro M. A., Pawlek B., Behrens B., Trautner T. A. Restriction and modification in Bacillus subtilis: expression of the cloned methyltransferase gene from B. subtilis phage SPR in E. coli and B. subtilis. Mol Gen Genet. 1983;189(1):17–20. doi: 10.1007/BF00326049. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Resistance to In Vitro Restriction of DNA from Lactic Streptococcal Bacteriophage c6A. Appl Environ Microbiol. 1986 Jun;51(6):1358–1360. doi: 10.1128/aem.51.6.1358-1360.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D. N., Eberle H., Bickle T. A. Characterization of mutations of the bacteriophage P1 mod gene encoding the recognition subunit of the EcoP1 restriction and modification system. J Bacteriol. 1989 May;171(5):2347–2352. doi: 10.1128/jb.171.5.2347-2352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988 Jan 11;16(1):355–355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Kita K., Takanami M. The FokI restriction-modification system. II. Presence of two domains in FokI methylase responsible for modification of different DNA strands. J Biol Chem. 1989 Apr 5;264(10):5757–5761. [PubMed] [Google Scholar]
- Tao T., Walter J., Brennan K. J., Cotterman M. M., Blumenthal R. M. Sequence, internal homology and high-level expression of the gene for a DNA-(cytosine N4)-methyltransferase, M.Pvu II. Nucleic Acids Res. 1989 Jun 12;17(11):4161–4175. doi: 10.1093/nar/17.11.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



