Abstract
Diacylglycerol can function as a second messenger, and one mechanism for the attenuation of this signal is its conversion to phosphatidic acid, which is catalyzed by diacylglycerol kinase (DGK). We screened a cDNA library from human skeletal muscle and isolated two DGKζ cDNAs that differed from the 3.5-kb clone originally identified in endothelial cells. One transcript, which was 3.4 kb long, was shown to be nonfunctional; it had a 77-bp deletion that included the translation initiation site. The other was 4.1 kb long with a unique 5′ sequence of 853 bp. We also isolated a genomic clone of DGKζ and determined its organization and location; it contains 32 exons, spans approximately 50 kb of genomic sequence, and maps to chromosome 11p11.2. The protein encoded by the 4.1-kb transcript contains two cysteine-rich regions, a catalytic domain, and ankyrin repeats like the endothelial form of DGKζ, as well as a unique N-terminal domain. The coding sequence was shown to be derived from alternative splicing of the DGKζ gene. In cells transfected with the 4.1-kb clone, we detected a 130-kDa protein with an antibody to DGKζ and demonstrated that it was localized predominantly in the nucleus. We conclude that alternative splicing generates tissue-specific variants of DGKζ that share some properties but may have unique ones as well.
In stimulated cells, the concentration of diacylglycerol (DAG) rises rapidly, and DAG functions as a second messenger by activating protein kinase C, which in turn regulates many cellular responses, including growth and differentiation (1). The mechanisms by which the level of DAG is regulated are not completely understood, and one unresolved issue is how the signaling function of DAG is distinguished from DAG’s central role in the biosynthesis of phospholipids and triglycerides. In both, the attenuation of the DAG signal and phospholipid synthesis, the conversion of DAG to phosphatidic acid (PA) is catalyzed by DAG kinases (DGKs) (2). The regulation of this reaction may be crucial because the product, PA, can also serve as a lipid messenger (3–5) and the net effect of conversion of DAG to PA might vary from cell to cell and condition to condition. Multiple isoforms of DGK, designated as DGKα (6, 7), DGKβ (8), DGKγ (9), DGKδ (10), DGKɛ (11), DGKζ (12, 13), and DGKη (14), have been identified by various approaches, and these isoforms differ in their Ca2+ dependency, substrate specificities, and structural domains.
Human DGKζ contains two cysteine-rich regions, a conserved catalytic domain with a presumed ATP-binding site, and four ankyrin repeats at the C terminus. A Northern blot of RNA from multiple human tissues showed the major transcript of DGKζ has an apparent size of 3.7 kb. However, an additional signal at an apparent size of 4.2 kb was detected in samples from skeletal and heart muscle (12). This result suggested that these muscles have an alternatively spliced form of DGKζ or another closely related isoform. Alternative splicing is an important means for the generation of protein diversity in eukaryotes. The alternative selection of exon sequences during splicing often is regulated in a developmental or a tissue-specific manner. A striking example is the Troponin T gene; five of its exons near the 5′ end are differentially incorporated in a developmental and tissue-specific manner to yield as many as 64 isoforms (15).
To further characterize DGKζ, we determined the genomic organization and chromosomal localization of DGKζ, and isolated and characterized two new cDNAs from muscle. Our results show that these muscle-specific DGKζ transcripts result from alternative splicing.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP (6,000 Ci/mmol; 1 Ci = 37 GBq), [α-32P]dCTP (6,000 Ci/mmol), enhanced chemiluminescence (ECL) detection reagents, and Hybond-N nylon membrane were purchased from Amersham. A skeletal muscle 5′-STRETCH cDNA library (λgt11, oligo(dT)+random primed) and human multiple-muscle Northern blot were purchased from CLONTECH. DMEM, penicillin, streptomycin, and Lipofectamine were from GIBCO/BRL, and fetal bovine serum was from HyClone. Leupeptin, pepstatin, aprotinin, and soybean trypsin inhibitor were purchased from Boehringer Mannheim. Avanti Polar Lipids provided phosphatidylserine and PA; all other lipids were from Serdary Research Laboratories (Englewood Cliffs, NJ). Octyl-β-glucopyranoside (ULTROL Grade) was purchased from Calbiochem. ATP and phenylmethylsulfonyl fluoride (PMSF) were purchased from Sigma. 4′,6-Diamidino-2-phenylindole was from Molecular Probes. Horseradish peroxidase and fluorescein-conjugated goat F(ab′)2 anti-rabbit immunoglobulin antibody were from Biosource International (Camarillo, CA). Prestained high molecular weight markers were from Bio-Rad.
Isolation of P1 Genomic Clone of DGKζ.
Replica membranes of a P1 human genomic library (University of Utah Genomics Core Facility) were screened with a 2-kb HindIII fragment of the human DGKζ cDNA that had been labeled with [α-32P]dCTP by hexanucleotide random priming. Membranes were prehybridized (4 h; 42°C) in 5× standard saline citrate (SSC)/10× Denhardt’s solution/0.05 M sodium phosphate, pH 6.7/500 μg/ml denatured human DNA/5% Dextran/50% formamide/0.5% SDS. Hybridization was performed (overnight; 42°C) in the same solution. The filters were washed once in 2× SSC/0.1% SDS and then in 0.1× SSC/0.1% SDS (20 min each; both at room temperature). Membranes were exposed to x-ray film overnight. After primary and secondary screening, three positive clones were isolated.
Mapping of Genomic Clones and Analysis of Exon/Intron Organization.
The positive P1 clones were analyzed by a combination of restriction mapping and Southern blot analysis. P1 DNA samples were digested with various restriction enzymes, separated on agarose gels, and analyzed by Southern blot hybridization using cDNA probes. Strongly positive bands were subcloned into a pBluescript II SK vector (Stratagene) and sequenced by automated dideoxy sequencing (Applied Biosystems, University of Utah DNA Sequencing Core Facility). A small segment of the cDNA sequence was not present in any of the subclones, and the PCR was performed to amplify two genomic fragments containing the missing sequence. The PCR products were subcloned into a pBluescript II SK vector for sequencing analysis. The PCR primers were 5′-GTGCAAAGATCATCCAGTCTT-3′, 5′-TGGAGAGGATCCAGCCCACCG-3′, 5′-ACCCCAATCCCTCTTTCCCAG-3′, and 5′-GAAAGACTGGATGATCTTTGC-3′. The DGKζ gene was physically mapped by fluorescence in situ hybridization using P137F4 DNA as a probe (16).
Isolation and Characterization of cDNAs.
One million phage recombinants were plated from a human skeletal muscle cDNA library. All plaques were transferred to Hybond-N nylon membranes (Amersham), which were screened with the previously described DGKζ cDNA (12). Membranes were prehybridized (4 h; 65°C) in 5× standard saline phosphate/EDTA (SSPE)/5× Denhardt’s solution/0.2% SDS/0.1% Na2P4O7. Hybridization was performed (overnight; 65°C) in the same solution. The membranes were washed twice in 0.6× SSPE/0.1% SDS/0.1% Na2P4O7 (15 min; 65°C). After primary and secondary screening, 18 positive clones were identified; all were analyzed by restriction digestion, and those of >3 kb were sequenced.
Cell Culture, Transfection, and DGK Activity Assay.
COS-7 cells were maintained in DMEM (GIBCO/BRL) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The two alternatively spliced forms were subcloned into pcDNAI/AMP (Invitrogen) in the forward orientation. COS-7 cells at 50–60% confluence in p35 dishes were transfected with 1 μg of plasmid DNA and 5 μl of Lipofectamine for 5 h according to the manufacturer’s guidelines. After 48 h, the cells were washed with PBS and were scraped into lysis buffer (20 mM Tris⋅HCl, pH 7.5/0.25 M sucrose/1 mM EDTA/4 mM EGTA/1 mM DTT/1 mM PMSF/20 μg/ml leupeptin, pepstatin, aprotinin, and soybean trypsin inhibitor). All homogenates were frozen and stored at −70°C until assayed; the assay was performed as described (12).
Western Blotting and Immunocytochemistry.
Samples from transfected cells were separated in a SDS/7.5% polyacrylamide gel, and Western blotting was performed as described (12). For immunocytochemical analysis, COS-7 cells were transfected with the alternatively spliced forms of DGKζ as described above. In the following procedure, the cells were washed three times with PBS between steps. After 48 h, the cells were fixed with 3.7% paraformaldehyde (10 min) and then permeabilized with 100% methanol (5 min; room temperature). The cells subsequently were blocked in PBS with 2% BSA and 2% goat serum (15 min; room temperature) and incubated with the anti-peptide DGKζ antibody (1:100 dilution) in blocking solution (30 min; 37°C). The cells were then incubated with a fluorescein-conjugated goat anti-rabbit immunoglobulin antibody (1:20 dilution) in blocking solution (30 min; 37°C) in the dark. The cells were counterstained with 4′,6-diamidino-2-phenylindole (1:5,000 dilution) for 1 min. The cells were photographed with fluorescent microscopy after mounting slides.
RESULTS
Isolation of a Muscle cDNA Encoding a Novel DGKζ.
A previous analysis of the distribution of DGKζ in human tissues showed that the predominant mRNA in most tissues was about 3.7 kb long but that skeletal and heart muscle had prominent bands at about 4.2 kb (12). We further examined DGKζ expression in muscles from different tissues and found the 4.1- to 4.2-kb mRNA in skeletal and heart muscle again, and also observed that it was present in smooth muscle from bladder and small intestine (Fig. 1). From the previous Northern blot (12) and this experiment, we conclude that the generation of the 4.1- to 4.2-kb transcript is muscle-specific and the levels of expression vary in different types of muscles. In addition, a band of approximately 9 kb was observed in skeletal and heart muscle, as reported previously (12). Our data suggest that this 9-kb transcript could be produced by incomplete splicing (data not shown).
Figure 1.
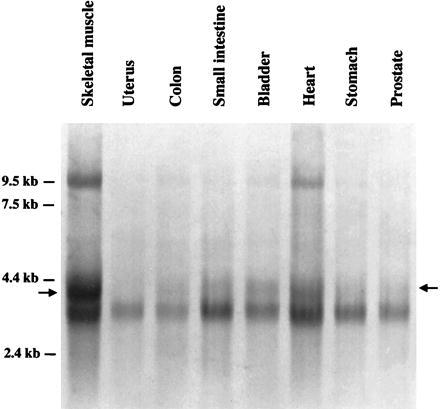
Analysis of DGKζ mRNA in muscle from human tissues. A filter with mRNA from muscle from various human tissues was probed with a fragment of the endothelial cDNA encoding DGKζ (12). All of the samples show a band at ≈3.7 kb as described (12). In addition, there was a prominent band at 4.1–4.2 kb (denoted with arrows) in the sample from skeletal muscle. Cardiac muscle and the muscle component from bladder and small intestine showed smaller amounts of this mRNA.
The 4.1- to 4.2-kb transcript detected in muscle could have been either an alternatively spliced form of DGKζ or a closely related, but different, isoform. To address this question, we screened a human skeletal muscle cDNA library and identified 18 positive clones; three of the clones shared a 5′ sequence that was different from the DGKζ cDNA isolated from an endothelial cell library (12). One of the three, which was about 4.1 kb, was analyzed by restriction digestion and fully sequenced. The clone had an 853-bp-long 5′ sequence that differed from the 249-bp-long 5′ sequence of the endothelial cell cDNA, including the translation initiation site (Fig. 2A). The 4.1-kb clone has a different predicted translation initiation site at position 124–126, which conforms well to the Kozak consensus sequence (Fig. 2B) (17). The muscle cDNA contained a single large ORF encoding a 1,117-aa protein with a calculated molecular mass of 124 kDa. From this initiation site, the 4.1-kb transcript would produce a protein with a unique N-terminal 242-aa sequence that does not show significant homology to any sequences in the existing databases. Apart from this 5′ region, the rest of the 4.1-kb clone had the same sequence as the DGKζ isolated from endothelial cells, including the polyadenylylation site. This finding suggested that alternative splicing was the mechanism by which this transcript was generated (see below).
Figure 2.
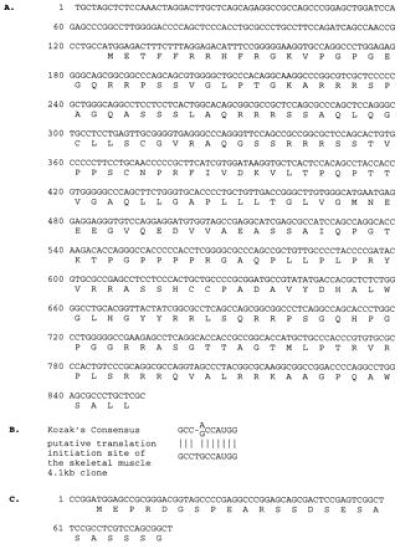
Sequence of DGKζ cDNAs isolated from skeletal muscle. A skeletal muscle library was screened with a probe from the endothelial cell DGKζ cDNA, and novel 4.1- and 3.4-kb cDNAs were isolated and sequenced. The 4.1-kb clone had a novel 5′ sequence, as compared with the endothelial cell clone, but was identical after position 853. The 3.4-kb clone was the same as that from endothelial cells except for a 77-bp deletion. (A) The unique nucleic acid sequence and deduced amino acid sequence of the 4.1-kb skeletal muscle clone. (B) A comparison between the putative translation initiation site of the 4.1-kb clone and the Kozak consensus sequence. (C) The deleted nucleotide sequence and corresponding amino acid sequence of the 3.4-kb clone.
In the screen of the muscle library, we isolated two additional clones (3.4 kb) that have the same 5′ sequence as the 3.5-kb cDNA from endothelial cells but contain a 77-bp deletion that includes the translation initiation site (Fig. 2C). This is consistent with the result of the previous Northern blot, which showed that an additional band in skeletal and heart muscle was slightly smaller than the major form in other tissues (12). Because of the deletion, this transcript cannot be translated into a functional DGK protein. Further, there is an in-frame stop codon upstream of the deleted ATG initiation site, a fact that excludes the possibility that an in-frame ATG site located further 5′ could be used.
Structure and Location of the Human DGKζ Gene.
We identified three positive clones in a human genomic library that reacted with the DGKζ cDNA from human endothelial cells. Based on Southern blot and sequence analyses, we concluded that one clone (P137F4), which was about 100 kb long, contained the entire coding sequence and substantial additional sequence, and we analyzed it further. The positions of exon-containing genomic fragments were determined by a combination of restriction endonuclease mapping and Southern blot hybridization, and the physical map and genomic organization of the human DGKζ gene are illustrated in Fig. 3. The exon/intron boundaries were determined by DNA sequence analyses of BamHI fragments that hybridized to a DGKζ cDNA probe (Table 1). The small gap between the 0.7- and 0.4-kb BamHI fragments was amplified by PCR from clone P137F4, subcloned, and sequenced. Using this analysis, we found that the coding sequence spanned about 50 kb and had 31 exons ranging in size from 32 to 732 bp. However, when we analyzed the sequence of the cDNA from the muscle clone, we found that the unique 5′ sequence was generated from an additional exon that lies between the previously determined first two exons. Thus, the gene has 32 exons; the previously described common form of DGKζ uses exons 1 and 3–32, whereas the muscle form uses exons 2–32 (Fig. 3).
Figure 3.
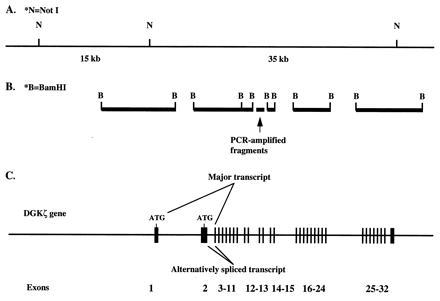
Organization of the human DGKζ gene. A genomic fragment of ≈100 kb was isolated and shown to contain the entire coding region for DGKζ. (A) The positions of NotI sites (indicated by N) are shown. (B) The sizes and positions of BamHI fragments that hybridized with the endothelial DGKζ cDNA are shown. (C) The intron-exon structure of the gene and the alternative splicing that yields the two different forms are shown. The exons are represented by vertical bars and numbers.
Table 1.
Exon/intron boundaries of the human DGKζ gene
| Exon | Length, bp | cDNA location | Splice acceptor Splice donor |
|---|---|---|---|
| 1 | Unknown | …–161 | ...TCGGGCACAG/gtgaacgggg |
| 2 | 744 | −16*–728* | ctgtgcccaa/CAGCCAACCG...CCCTGCTCGC/ctaggtatag |
| 3 | 110 | 162–271 | tcctctctag/GAAAGCCATC...GACTGGAGCG/tgagtgcctg |
| 4 | 95 | 272–366 | cgtggctaga/AGTCAGCGAC...CAGGATGCTG/gtgagtgctc |
| 5 | 75 | 367–441 | cctgcagcag/AAGTCAGTGT...GCTGGAGAAG/gtgggtgggt |
| 6 | 57 | 442–498 | atgttcacag/ATAAATTTCC...TGTCCGCGAG/gtaagtgccc |
| 7 | 69 | 499–567 | tctcccccag/CCAACCTTTG...CTGTGGGAAG/gtgagaggcc |
| 8 | 72 | 568–639 | ctaccctcag/GGATTCCAGC...CAAGCAGGCA/gtgagtggtg |
| 9 | 117 | 640–756 | ctctcctcag/TACCACAGCA...GAGGCCCCAG/gtgagtactg |
| 10 | 72 | 757–828 | tcccttgcag/AATACTCTGA...AGGGCCTGAG/gtcagcccca |
| 11 | 96 | 829–924 | tgtctcccag/GAGGGCCGCT...GGGCAACCAG/gtgaacgcgg |
| 12 | 83 | 925–1,007 | tcccatccag/GGTGCAAAGA...CCAAGGAGGC/gtaagtactt |
| 13 | 61 | 1,008–1,068 | atgagcccag/GCTGGAGATG...CGACGGCACG/gtgagcttcc |
| 14 | 114 | 1,069–1,182 | cctccccccg/GTGGGCTGGA...CTGGGGTGGG/gtaagcaccc |
| 15 | 141 | 1,183–1,323 | gctcccccag/GGCTACACAG...CACCGACCGG/gtaagttggc |
| 16 | 79 | 1,324–1,402 | ctttttccag/TTGCCCCTGG...GAGTCTCGAG/gttggcagcc |
| 17 | 56 | 1,403–1,458 | ctccacccag/AGGCCAACCC...CTACGCCGGG/gtgagtgggg |
| 18 | 63 | 1,459–1,521 | tctcccacag/ACAGCTTTCT...CCGAGTGGTG/gtgagcgggg |
| 19 | 74 | 1,522–1,595 | ccgcctccag/TGTGATGGAA...ACATCCCCAG/gtgaggaggg |
| 20 | 112 | 1,596–1,707 | gcctttcaag/GTACTGTGCG...GACGTCGTTG/gtgagtgggc |
| 21 | 200 | 1,708–1,907 | gtgcccacag/GCCGCGCTGC...TGCACAGCGA/gtacgtccca |
| 22 | 101 | 1,908–2,008 | tgcccgacag/CCAGCAGCCG...AAGGAGGCCT/gtgagtgcgg |
| 23 | 80 | 2,009–2,088 | tgttctccag/CTGTGCCGCT...ACTCCAGCAG/gtaaggggtg |
| 24 | 70 | 2,089–2,158 | ctctcaacag/GAGCCCGATG...TTCCTGGACG/gtgagtctac |
| 25 | 41 | 2,159–2,199 | tttcccacag/CCACCACTGC...CCGAGCCCAG/gtgagcgatt |
| 26 | 140 | 2,200–2,339 | ccccccacag/GAGCACCTCA...CCACGCCCCG/gtgagtcctg |
| 27 | 32 | 2,340–2,371 | tctgttgcag/GTCACTGCAA...CCCCCTCAAG/gtgaggcctc |
| 28 | 44 | 2,372–2,415 | ttccgtgcag/GTGAAGAGCT...CTTCTGTAAG/gtactagctg |
| 29 | 121 | 2,416–2,536 | ctgccctcag/CTCCAGGAGC...CTGGACCACG/gtgagccggg |
| 30 | 34 | 2,537–2,570 | ccttctccag/CCCCCCCAGA...TGGAGGAAAA/gtaagtatct |
| 31 | 100 | 2,571–2,670 | gctcccccag/CGGGGAGACC...AGACCAGCAG/gtgagcagac |
| 32 | 732 | 2,671–3,402 | cctcctccag/GGCGACACTC...Polyadenylylation |
Nucleotide sequences around the exon/intron boundaries are presented. Exon sequences are shown in uppercase letters, and intron sequences are shown in lowercase letters. The locations of the splice junctions are indicated by slashes. The first base of the ATG initiation codon is designated +1.
Positions in the 4.1-kb muscle cDNA.
To determine the physical location of the DGKζ gene, we conducted fluorescence in situ hybridization with clone P137F4 and found that it localized to chromosome 11p11.2 (data not shown).
The Muscle-Specific DGKζ cDNA Encodes a Functional Enzyme Located in the Nucleus.
To further analyze the 4.1- and 3.4-kb cDNAs isolated from skeletal muscle, we subcloned each of them into an expression vector and transfected them into COS-7 cells. Proteins with apparent molecular masses of 114 and 130 kDa were recognized by an antibody to DGKζ in the cells transfected with the endothelial DGKζ cDNA and the skeletal muscle 4.1-kb cDNA, respectively. The molecular mass of the protein expressed from the 4.1-kb cDNA compares favorably with the predicted size of 124 kDa. Extracts from cells transfected with vector alone or with the 3.4-kb cDNA from skeletal muscle did not react strongly with the antibody (Fig. 4). Further, preincubation of the antibody with the peptide antigen blocked the recognition of the 114- and 130-kDa proteins, confirming that the interaction between the antibody and these proteins was specific. Thus, the larger, but not the smaller, clone from skeletal muscle was translated.
Figure 4.
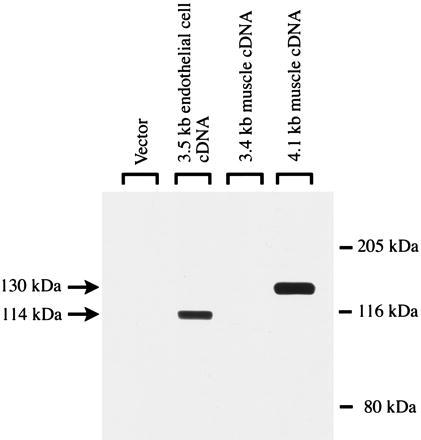
The 4.1-kb cDNA from skeletal muscle is expressed as a 130-kDa protein. COS-7 cells were transfected with the two DGKζ cDNAs isolated from skeletal muscle (the 4.1-kb alternatively spliced form and the 3.4-kb form with a deletion), the endothelial cell cDNA, or vector alone. After 48 h, the cells were harvested and examined by Western blotting for expression of DGKζ with a previously described antibody (12). Forty micrograms of protein of the COS-7 cell homogenate was loaded in each lane. In control experiments, preincubation with the immunogen-peptide significantly blocked the recognition of the 114- and 130-kDa proteins.
Homogenates from cells transfected with the muscle 4.1-kb cDNA, vector alone, or the endothelial cell cDNA also were assayed for DGK activity. The DGK activity in cells transfected with 4.1-kb clone was more than 50-fold higher than those with vector alone and similar to those transfected with the endothelial cell cDNA. We also compared the substrate selectivity of the skeletal muscle form with the endothelial DGKζ (Fig. 5). Both phosphorylated 1,2-diacylglycerols, but not 1,3-diacylglycerols, demonstrated greater activity in the presence of short chain, as compared with long chain, DAGs. However, there were no substantial differences in their use of other DAGs.
Figure 5.
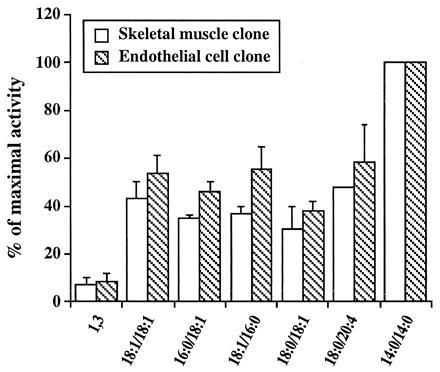
Substrate use by the activities expressed from the major and alternatively spliced forms of DGKζ. COS-7 cells were transfected with vectors encoding the 4.1-kb (skeletal muscle) cDNA and the 3.5-kb (endothelial cell) cDNA, respectively, and the homogenates were assayed for DGK activity as described in Material and Methods. The total activity varied among experiments, but in each one, the homogenates from cells transfected with the two different cDNAs were assayed identically in parallel. In a typical experiment, the DGK activity was 7.5 and 6.8 nmol/mg/min of protein for the endothelial cell and muscle forms, respectively, using 1,2-dicapryl-sn-glycerol as the substrate. The results are presented as the percentage of the activity observed against 1,2-dicapryl-sn-glycerol substrate (which is reported as 100% for both forms) and represent the mean of the values obtained in two separate experiments. The error bars indicate the standard deviation. 18:1/18:1, 1,2-dioleoyl-sn-glycerol; 1,3, 1,3-dioleoyl-sn-glycerol; 16:0/18:1, 1-palmitoyl-2-oleoyl-sn-glycerol; 18:1/16:0, 1-oleoyl-2-palmitoyl-sn-glycerol; 18:0/18:1, 1-stearoyl-2-oleoyl-sn-glycerol; 18:0/20:4, 1-stearoyl-2-arachidonyl-sn-glycerol; 14:0/14:0, 1,2-dicapryl-sn-glycerol.
In recent experiments, we found that the endothelial form of DGKζ is expressed in the cell nucleus (M.K.T., M.B., G.A.Z., T.M.M., P. J. Blackshew, and S.M.P., unpublished work), and we tested whether the alternatively spliced form from muscle also is targeted there. As shown in Fig. 6, the immunoreactive protein in DGKζ-transfected cells colocalized with the counterstain for DNA, establishing a nuclear location. Again, inclusion of the peptide to which the antibody had been raised completely abolished the nuclear staining.
Figure 6.
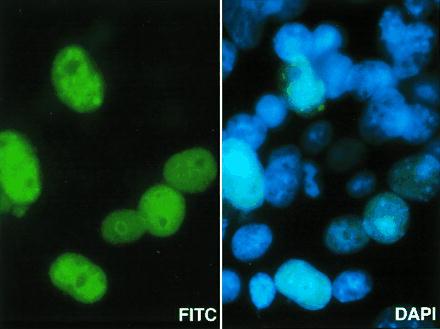
The alternatively spliced form of DGKζ is expressed in the cell nucleus. COS-7 cells were transfected with the 4.1-kb skeletal muscle DGKζ cDNA for 48 h and then were fixed with paraformaldehyde. They were stained with a primary antibody to DGKζ, which was detected with a secondary fluorescein-conjugated antibody and examined by fluorescence microscopy. The green staining shows the location of DGKζ in cells that had been transfected successfully. Control experiments in which the primary antibody was omitted or preincubated with the immunogen-peptide all showed no staining for DGKζ. The cells also were counterstained with 4′,6-diamidino-2-phenylindole to identify DNA. The same field is shown in both panels; the blue staining identifies the nucleus of all the cells in the field. (×1,000.)
DISCUSSION
The conversion of DAG to PA may be an important mechanism to attenuate the second messenger function of DAG. Alternatively, the PA produced by this reaction might have signaling functions itself. Because both of these lipids have important roles in complex lipid synthesis, this reaction may regulate that cellular function as well. Recently, several isoforms of DGK have been cloned and studied, but many issues remain. Are there multiple forms in each cell or is there tissue and cell specificity? What function(s) are served by so many isoforms? How are the signaling and membrane synthesis pathways isolated (or are they)? To address these and other topics, we isolated a cDNA encoding an isoform, designated ζ, which has interesting structural properties: a region homologous to the myristoylated alanine-rich C kinase substrate phosphorylation site domain and multiple ankyrin repeats, in addition to the cysteine-rich regions and a catalytic region found in all of the DGK isoforms (12). In recent work, we have shown that the myristoylated alanine-rich C kinase substrate sequence is a nuclear localization signal and that it is regulated by phosphorylation (Topham et al., unpublished work).
In our earlier studies, we noted that there was a messenger RNA in muscle that hybridized with the DGKζ cDNA but had an apparent size of 4.1 kb rather than 3.5 kb (12). This larger form was the major band in muscle samples. In the work reported here, we showed that this mRNA is expressed in other muscles, and we isolated its cDNA from a skeletal muscle library. To address the issue of the source of this cDNA, we also isolated a genomic clone and determined the gene structure. From this experiment, we showed that the muscle mRNA is derived from alternative splicing of the gene. A previous study suggested that human DGKζ might map to chromosomes 2 or 11 (18). Our result demonstrated that the DGKζ gene is located on chromosome 11p11.2, contains 32 exons, and spans about 50 kb of genomic sequence. We previously noted that portions of the DGKζ coding region were homologous to the rdgA gene of Drosophila (12), and by determining the exon/intron boundaries of the DGKζ gene, we found that some of the splice junctions also are well conserved (19). Interestingly, we also found the 3′ untranslated region of DGKζ is identical to the promoter region of heparin-binding cytokine midkine which has been mapped to human chromosome 11p13–11 (20).
We found two alternatively spliced DGKζ transcripts in a human skeletal muscle cDNA library; one is 4.1 kb long with a unique 5′ coding sequence and represents the previously recognized major form in muscle (12). It is expressed because a protein with an apparent molecular mass of 130 kDa was detected by an antibody to a peptide from DGKζ when COS-7 cells were transfected with the 4.1-kb cDNA clone. Moreover, the DGK activity of the cells rose markedly, and the use of different DAGs was like that of the activity derived form the 3.5-kb clone, which was examined in parallel. As with the 3.5-kb clone, most of the 4.1-kb DGKζ was found in the nucleus of transfected cells. We also isolated a 3.4-kb clone with a 77-bp deletion that included the translation initiation site. This transcript did not encode a DGKζ protein.
By analysis of the genomic structure and cDNA sequence comparisons, we found that the cDNAs isolated from endothelial cells and skeletal muscle are derived from the same gene but with different exon use. The basis for this is likely to be tissue-specific factors that regulate gene expression. Similar alternative splicing has been found in the prolactin receptor genes and, in that situation, multiple tissue-specific promoters yield different expression in gonadal versus non-gonadal tissues (21). In muscle, specific trans-acting factors that are developmentally related have been reported (15), and a related mechanism could explain some of our findings because in other experiments we have observed a marked change in the expression of mouse DGKζ during embryogenesis. We speculate that alternative splicing in DGKζ is differentially regulated in different muscle cell types and possibly in different developmental stages of the same muscle lineage. Interestingly, alternative splicing was also observed in DGKγ (9); functional DGKγ is predominantly expressed in human retina, but a truncated, inactive form containing a 25-aa deletion within the catalytic domain was identified in many human cells. Therefore, alternative splicing may be a common mechanism in regulating DGK activity in different tissues. However, the utility of this form of regulation is not yet clear because in the case of DGKγ the alternative form was without catalytic function. In our studies, the alternatively spliced DGKζ was functional, but its properties were essentially the same as for the more widely expressed form. However, DGKζ activity may be affected in vivo by cofactors and relative substrate abundance, which could differ between muscle and other tissues. Also, the different primary structure may confer subtle effects that were not detected in our assays.
Acknowledgments
We thank Elie Traer for assistance with the computer searches and for the figures reporting sequence analysis. This work was supported by National Cancer Institute Grant CA59548. The core facilities at the University of Utah (DNA Sequencing, Genomics, Cytogenetics, and Peptide/DNA User Facility) all are supported by Grant CA42014 from the National Cancer Institute.
ABBREVIATIONS
- DAG
diacylglycerol
- PA
phosphatidic acid
- DGK
DAG kinase
Footnotes
References
- 1.Kikkawa U, Kishimoto A, Nishizuka Y. Annu Rev Biochem. 1989;58:31–44. doi: 10.1146/annurev.bi.58.070189.000335. [DOI] [PubMed] [Google Scholar]
- 2.Kanoh H, Yamada K, Sakane F. Trends Biochem Sci. 1990;15:47–50. doi: 10.1016/0968-0004(90)90172-8. [DOI] [PubMed] [Google Scholar]
- 3.Moolenaar W H, Kruijer W, Tilly B C, Verlaan I, Bierman A J, de Laat S W. Nature (London) 1986;323:171–173. doi: 10.1038/323171a0. [DOI] [PubMed] [Google Scholar]
- 4.Knauss T C, Jaffer F E, Abboud H E. J Biol Chem. 1990;265:14457–14463. [PubMed] [Google Scholar]
- 5.Fukami K, Takenawa T. J Biol Chem. 1992;267:10988–10993. [PubMed] [Google Scholar]
- 6.Sakane F, Yamada K, Kanoh H, Yokoyama C, Tanabe T. Nature (London) 1990;344:345–348. doi: 10.1038/344345a0. [DOI] [PubMed] [Google Scholar]
- 7.Schaap D, de Widt J, van der Wal J, Vandekerckhove J, van Damme J, Gussow D, Ploegh H L, van Blitterswijk W J, van der Bend R L. FEBS Lett. 1990;275:151–158. doi: 10.1016/0014-5793(90)81461-v. [DOI] [PubMed] [Google Scholar]
- 8.Goto K, Kondo H. Proc Natl Acad Sci USA. 1993;90:7598–7602. doi: 10.1073/pnas.90.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kai M, Sakane F, Imai S, Wada I, Kanoh H. J Biol Chem. 1994;269:18492–18498. [PubMed] [Google Scholar]
- 10.Sakane F, Imai S, Kai M, Wada I, Kanoh H. J Biol Chem. 1996;271:8394–8401. doi: 10.1074/jbc.271.14.8394. [DOI] [PubMed] [Google Scholar]
- 11.Tang W, Bunting M, Zimmerman G A, McIntyre T M, Prescott S M. J Biol Chem. 1996;271:10237–10241. [PubMed] [Google Scholar]
- 12.Bunting M, Tang W, Zimmerman G A, McIntyre T M, Prescott S M. J Biol Chem. 1996;271:10230–10236. [PubMed] [Google Scholar]
- 13.Goto K, Kondo H. Proc Natl Acad Sci USA. 1996;93:11196–11201. doi: 10.1073/pnas.93.20.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klauck T M, Xu X, Mousseau B, Jaken S. J Biol Chem. 1996;271:19781–19788. doi: 10.1074/jbc.271.33.19781. [DOI] [PubMed] [Google Scholar]
- 15.Breitbart R E, Nadal-Ginard B. Cell. 1987;49:793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- 16.Pinkel D, Straume T, Gray J W. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilz A, Schaap D, Hunt D, Fitzgibbon J. Genomics. 1995;26:599–601. doi: 10.1016/0888-7543(95)80182-l. [DOI] [PubMed] [Google Scholar]
- 19.Masai I, Okazaki A, Hosoya T, Hotta Y. Proc Natl Acad Sci USA. 1993;90:11157–11161. doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara B, Jenkins N A, Gilbert D J, Copeland N G, Shows T B, Eddy R L, Bohlen P, Kovesdi I. Cytogenet Cell Genet. 1995;69:40–43. doi: 10.1159/000133934. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, Zhuang L, Dufau M L. J Biol Chem. 1996;271:10242–10246. doi: 10.1074/jbc.271.17.10242. [DOI] [PubMed] [Google Scholar]


