Abstract
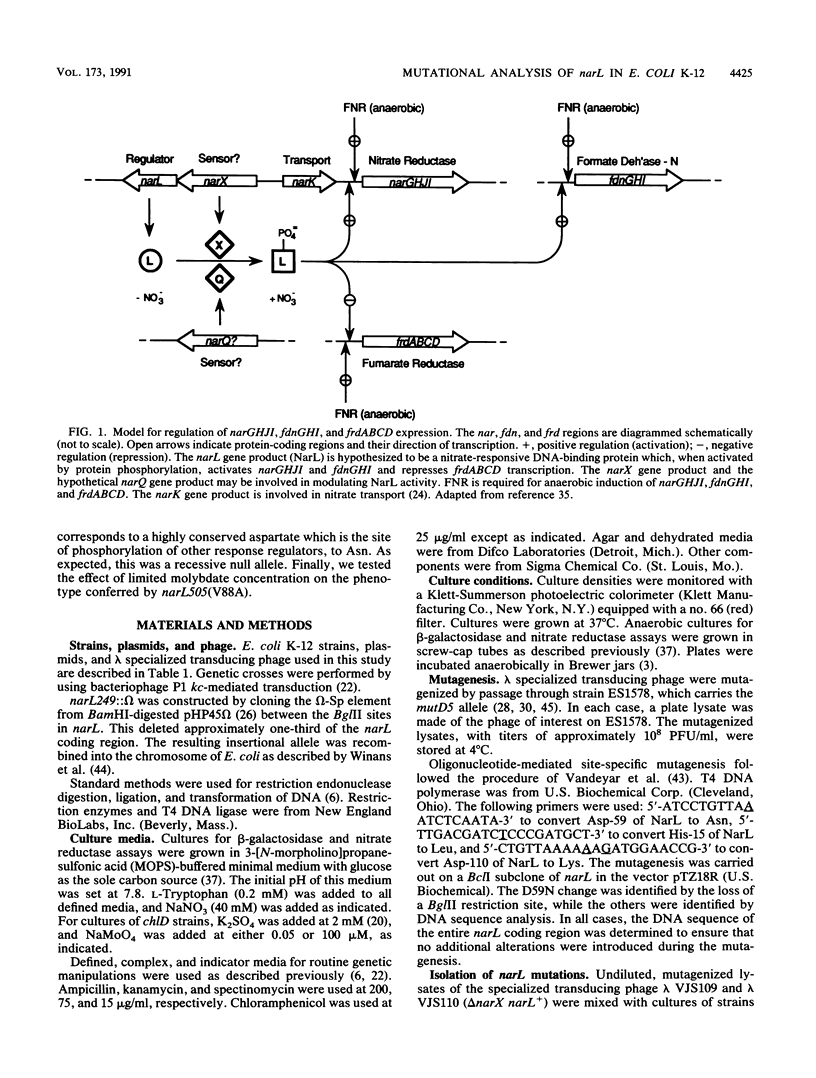
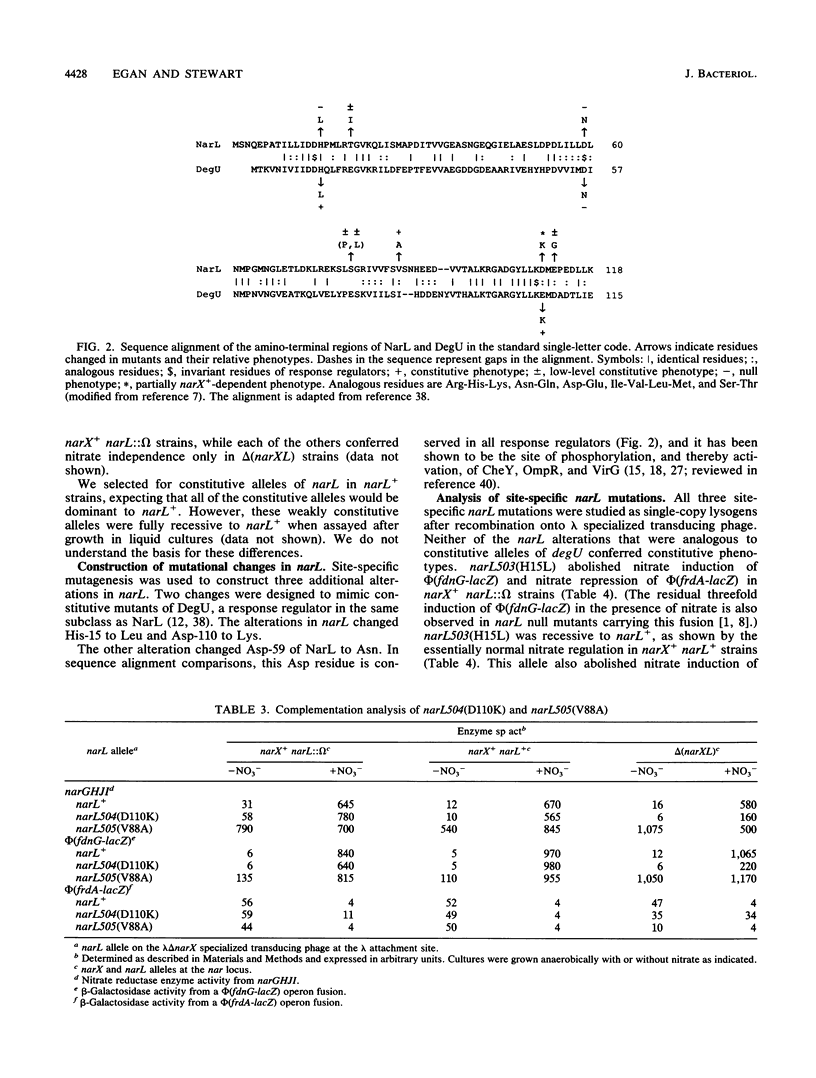
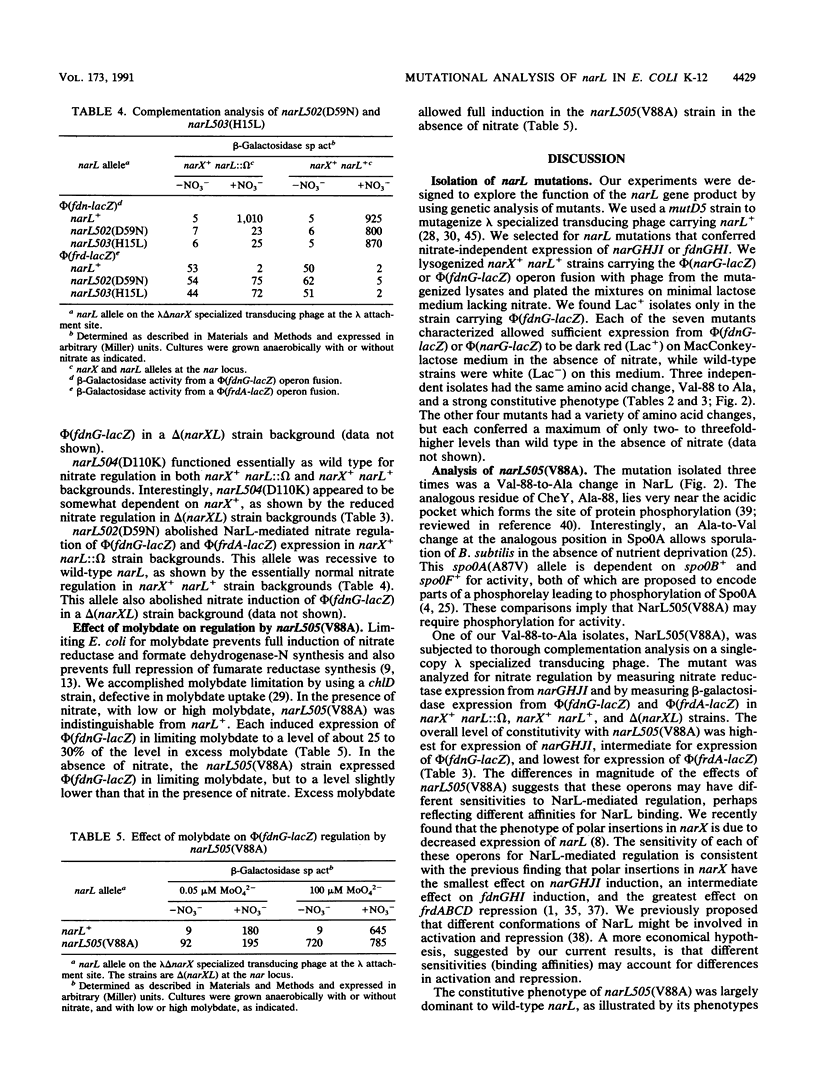
The narL gene product, NarL, is the nitrate-responsive regulator of anaerobic respiratory gene expression. We used genetic analysis of narL mutants to better understand the mechanism of NarL-mediated gene regulation. We selected and analyzed seven nitrate-independent narL mutants. Each of three independent, strongly constitutive mutants had changes of Val-88 to Ala. The other four mutants were weakly constitutive. The narL505(V88A) allele was largely dominant to narL+, while narX+ had a negative influence on its constitutive phenotype, suggesting that NarX may play a negative role in nitrate regulation. We also constructed two narL mutations that are analogous to previously characterized constitutive degU alleles. The first, narL503(H15L), was a recessive null allele. The second, narL504(D110K), functioned essentially as wild type but was dependent on narX+ for full activity. We changed Asp-59 of NarL, which corresponds to the site of phosphorylation of other response regulators, to Asn. This change, narL502(D59N), was a recessive null allele, which is consistent with the hypothesis that NarL requires phosphorylation for activation. Finally, we tested the requirement for molybdate on regulation in a narL505(V88A) strain. Although narL505(V88A) conferred some nitrate-independent expression of fdnGHI (encoding formate dehydrogenase-N) in limiting molybdate, it required excess molybdate for full induction both in the absence and in the presence of nitrate. This finding suggests that narL505(V88A) did not confer molybdate-independent expression of fdnGHI.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg B. L., Stewart V. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics. 1990 Aug;125(4):691–702. doi: 10.1093/genetics/125.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer J. H., Allgeier D. L. Safe Self-contained Carbon Dioxide-Hydrogen Anaerobic System. Appl Microbiol. 1966 Nov;14(6):985–988. doi: 10.1128/am.14.6.985-988.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Dahl M. K., Msadek T., Kunst F., Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991 Apr;173(8):2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. M., Stewart V. Nitrate regulation of anaerobic respiratory gene expression in narX deletion mutants of Escherichia coli K-12. J Bacteriol. 1990 Sep;172(9):5020–5029. doi: 10.1128/jb.172.9.5020-5029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Kalman L. V., Stewart R. R. Nucleotide sequence of the narL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res. 1989 Mar 11;17(5):1965–1975. doi: 10.1093/nar/17.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Yang M., Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. J Bacteriol. 1988 Nov;170(11):5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. Molybdenum effector of fumarate reductase repression and nitrate reductase induction in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3720–3725. doi: 10.1128/jb.169.8.3720-3725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G., Prusti R. K., Roitsch T., Ankenbauer R. G., Nester E. W. Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J Bacteriol. 1990 Sep;172(9):4945–4950. doi: 10.1128/jb.172.9.4945-4950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L. V., Gunsalus R. P. Identification of a second gene involved in global regulation of fumarate reductase and other nitrate-controlled genes for anaerobic respiration in Escherichia coli. J Bacteriol. 1989 Jul;171(7):3810–3816. doi: 10.1128/jb.171.7.3810-3816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L. V., Gunsalus R. P. Nitrate- and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J Bacteriol. 1990 Dec;172(12):7049–7056. doi: 10.1128/jb.172.12.7049-7056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K., Aiba H., Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: I. Analysis by site-directed mutagenesis of the amino acid residues involved in phosphotransfer between the two regulatory components, EnvZ and OmpR. J Biochem. 1990 Sep;108(3):483–487. doi: 10.1093/oxfordjournals.jbchem.a123225. [DOI] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Lee J. H., Wendt J. C., Shanmugam K. T. Identification of a new gene, molR, essential for utilization of molybdate by Escherichia coli. J Bacteriol. 1990 Apr;172(4):2079–2087. doi: 10.1128/jb.172.4.2079-2087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohno T., Noji S., Taniguchi S., Saito T. The narX and narL genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryotic two-component regulatory genes. Nucleic Acids Res. 1989 Apr 25;17(8):2947–2957. doi: 10.1093/nar/17.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji S., Nohno T., Saito T., Taniguchi S. The narK gene product participates in nitrate transport induced in Escherichia coli nitrate-respiring cells. FEBS Lett. 1989 Jul 31;252(1-2):139–143. doi: 10.1016/0014-5793(89)80906-8. [DOI] [PubMed] [Google Scholar]
- Olmedo G., Ninfa E. G., Stock J., Youngman P. Novel mutations that alter the regulation of sporulation in Bacillus subtilis. Evidence that phosphorylation of regulatory protein SpoOA controls the initiation of sporulation. J Mol Biol. 1990 Oct 5;215(3):359–372. doi: 10.1016/s0022-2836(05)80357-2. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Schaaper R. M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D., Amy N. K. Molybdenum accumulation in chlD mutants of Escherichia coli. J Bacteriol. 1989 Mar;171(3):1284–1287. doi: 10.1128/jb.171.3.1284-1287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Stewart G. S., Lubinsky-Mink S., Jackson C. G., Cassel A., Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986 May;15(3):172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Stewart V., Berg B. L. Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase gene expression in Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4437–4444. doi: 10.1128/jb.170.10.4437-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1988 Apr;170(4):1589–1597. doi: 10.1128/jb.170.4.1589-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr, Merkel S. M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1989 Apr;171(4):2229–2234. doi: 10.1128/jb.171.4.2229-2234.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A. M., Mottonen J. M., Stock J. B., Schutt C. E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989 Feb 23;337(6209):745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991 Mar;173(5):1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. H., Clarke C. H., Marinus M. G. Specificity of Escherichia coli mutD and mutL mutator strains. Gene. 1990 Mar 1;87(1):1–5. doi: 10.1016/0378-1119(90)90488-d. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



