Abstract
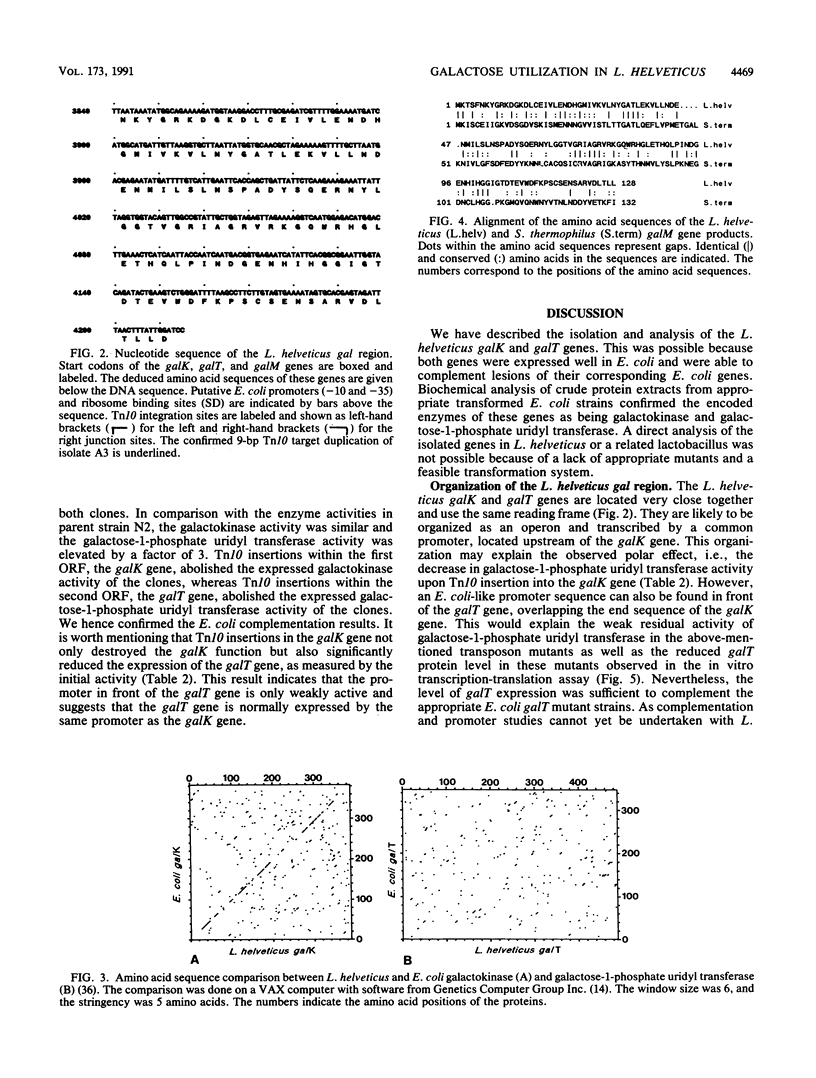
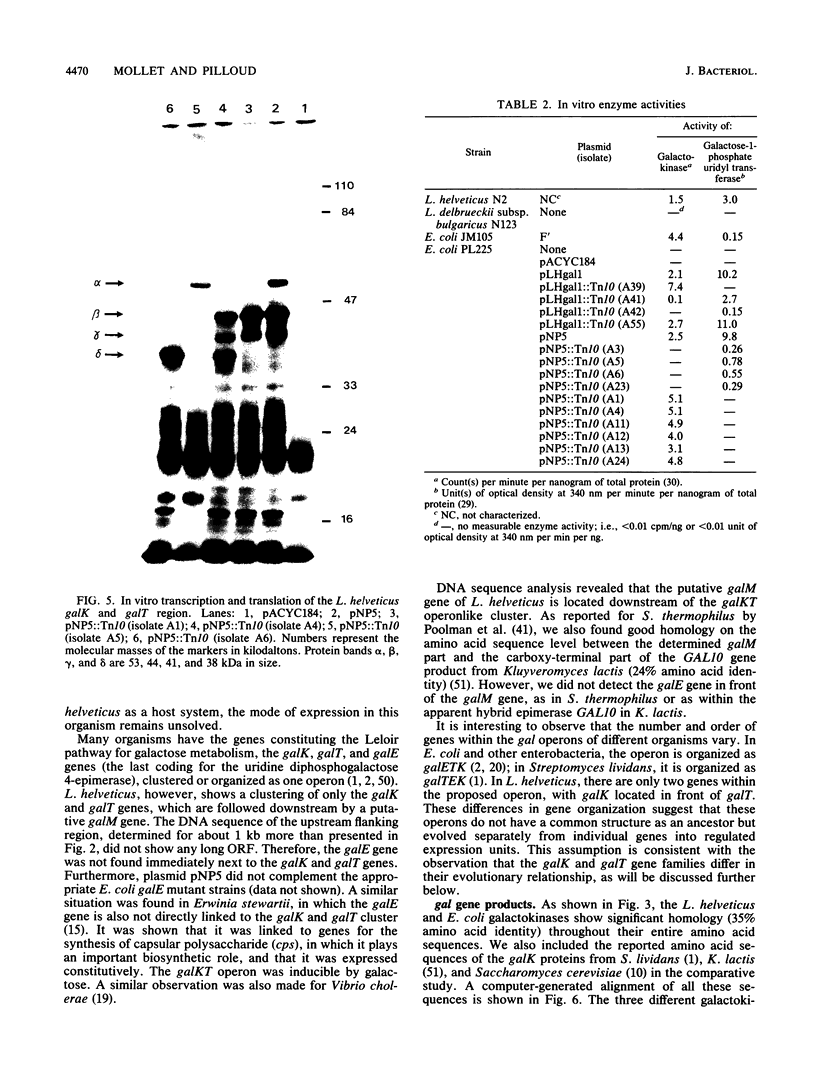
By complementing appropriate gal lesions in Escherichia coli K802, we were able to isolate the galactokinase (galK) and galactose-1-phosphate uridyl transferase (galT) genes of Lactobacillus helveticus. Tn10 transposon mutagenesis, together with in vivo complementation analysis and in vitro enzyme activity measurements, allowed us to map these two genes. The DNA sequences of the genes and the flanking regions were determined. These revealed that the two genes are organized in the order galK-galT in an operonlike structure. In an in vitro transcription-translation assay, the galK and galT gene products were identified as 44- and 53-kDa proteins, respectively, data which corresponded well with the DNA sequencing data. The deduced amino acid sequence of the galK gene product showed significant homologies to other prokaryotic and eukaryotic galactokinase sequences, whereas galactose-1-phosphate uridyl transferase did not show any sequence similarities to other known proteins. This observation, together with a comparison of known gal operon structures, suggested that the L. helveticus operon developed independently to a translational expression unit having a different gene order than that in E. coli, Streptococcus lividans, or Saccharomyces cerevisiae. DNA sequencing of the flanking regions revealed an open reading frame downstream of the galKT operon. It was tentatively identified as galM (mutarotase) on the basis of the significant amino acid sequence homology with the corresponding Streptococcus thermophilus gene.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Fornwald J. A., Schmidt F. J., Rosenberg M., Brawner M. E. Gene organization and structure of the Streptomyces lividans gal operon. J Bacteriol. 1988 Jan;170(1):203–212. doi: 10.1128/jb.170.1.203-212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Boizet B., Flickinger J. L., Chassy B. M. Transfection of Lactobacillus bulgaricus protoplasts by bacteriophage DNA. Appl Environ Microbiol. 1988 Dec;54(12):3014–3018. doi: 10.1128/aem.54.12.3014-3018.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B. A., Donelson J. E. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol. 1984 Apr;158(1):269–278. doi: 10.1128/jb.158.1.269-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck C., Riccio A., Schumperli D., McKenney K., Jeffers J., Hughes C., Rosenberg M., Heusterspreute M., Brunel F., Davison J. Structure of the galactokinase gene of Escherichia coli, the last (?) gene of the gal operon. Nucleic Acids Res. 1985 Mar 25;13(6):1841–1853. doi: 10.1093/nar/13.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley M., Mollet B., Hottinger H. DNA Probe for Lactobacillus delbrueckii. Appl Environ Microbiol. 1990 Jun;56(6):1967–1970. doi: 10.1128/aem.56.6.1967-1970.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph P. J., Majerczak D. R., Coplin D. L. Characterization of a gene cluster for exopolysaccharide biosynthesis and virulence in Erwinia stewartii. J Bacteriol. 1988 Feb;170(2):865–871. doi: 10.1128/jb.170.2.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Hashiba H., Takiguchi R., Ishii S., Aoyama K. Transformation of Lactobacillus helveticus subsp. jugurti with plasmid pLHR by electroporation. Agric Biol Chem. 1990 Jun;54(6):1537–1541. [PubMed] [Google Scholar]
- Hickey M. W., Hillier A. J., Jago G. R. Transport and metabolism of lactose, glucose, and galactose in homofermentative lactobacilli. Appl Environ Microbiol. 1986 Apr;51(4):825–831. doi: 10.1128/aem.51.4.825-831.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houng H. S., Kopecko D. J., Baron L. S. Molecular cloning and physical and functional characterization of the Salmonella typhimurium and Salmonella typhi galactose utilization operons. J Bacteriol. 1990 Aug;172(8):4392–4398. doi: 10.1128/jb.172.8.4392-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutkins R. W., Ponne C. Lactose Uptake Driven by Galactose Efflux in Streptococcus thermophilus: Evidence for a Galactose-Lactose Antiporter. Appl Environ Microbiol. 1991 Apr;57(4):941–944. doi: 10.1128/aem.57.4.941-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979 Apr;16(4):711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langella P., Chopin A. Conjugal transfer of plasmid pIP501 from Lactococcus lactis to Lactobacillus delbrückii subsp. bulgaricus and Lactobacillus helveticus. FEMS Microbiol Lett. 1989 Jul 15;51(1):149–152. doi: 10.1016/0378-1097(89)90498-9. [DOI] [PubMed] [Google Scholar]
- Lemaire H. G., Müller-Hill B. Nucleotide sequences of the gal E gene and the gal T gene of E. coli. Nucleic Acids Res. 1986 Oct 10;14(19):7705–7711. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Fukasawa T. Chromosome mobilization in rec-merodiploids of Escherichia coli K12 following infection with bacteriophage lambda. Virology. 1969 Nov;39(3):467–481. doi: 10.1016/0042-6822(69)90095-6. [DOI] [PubMed] [Google Scholar]
- Mollet B., Delley M. A beta-galactosidase deletion mutant of Lactobacillus bulgaricus reverts to generate an active enzyme by internal DNA sequence duplication. Mol Gen Genet. 1991 May;227(1):17–21. doi: 10.1007/BF00260700. [DOI] [PubMed] [Google Scholar]
- Mollet B., Delley M. Spontaneous deletion formation within the beta-galactosidase gene of Lactobacillus bulgaricus. J Bacteriol. 1990 Oct;172(10):5670–5676. doi: 10.1128/jb.172.10.5670-5676.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Novotny J. Matrix program to analyze primary structure homology. Nucleic Acids Res. 1982 Jan 11;10(1):127–131. doi: 10.1093/nar/10.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Shimada K., Tomizawa J. Studies on radiation-sensitive mutants of E. coli. I. Mutants defective in the repair synthesis. Mol Gen Genet. 1968 May 3;101(3):227–244. doi: 10.1007/BF00271625. [DOI] [PubMed] [Google Scholar]
- Poolman B. Precursor/product antiport in bacteria. Mol Microbiol. 1990 Oct;4(10):1629–1636. doi: 10.1111/j.1365-2958.1990.tb00539.x. [DOI] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990 Jul;172(7):4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989 Jan;171(1):244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt J. K., Berg P. Cloning and characterization of a cDNA encoding human galactose-1-phosphate uridyl transferase. Mol Biol Med. 1988 Apr;5(2):107–122. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tajima M., Nogi Y., Fukasawa T. Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast. 1985 Sep;1(1):67–77. doi: 10.1002/yea.320010108. [DOI] [PubMed] [Google Scholar]
- Thompson J. K., Collins M. A. Evidence for the conjugal transfer of the broad host range plasmid pIP501 into strains of Lactobacillus helveticus. J Appl Bacteriol. 1988 Oct;65(4):309–319. doi: 10.1111/j.1365-2672.1988.tb01897.x. [DOI] [PubMed] [Google Scholar]
- Turner K. W., Martley F. G. Galactose fermentation and classification of thermophilic lactobacilli. Appl Environ Microbiol. 1983 Jun;45(6):1932–1934. doi: 10.1128/aem.45.6.1932-1934.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Way J. C., Kleckner N. Essential sites at transposon Tn 10 termini. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3452–3456. doi: 10.1073/pnas.81.11.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T. D., Dickson R. C. Nucleotide sequence of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 1988 Aug 25;16(16):8192–8194. doi: 10.1093/nar/16.16.8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T. D., Dickson R. C. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis. Nucleic Acids Res. 1988 Aug 25;16(16):8011–8028. doi: 10.1093/nar/16.16.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



