Abstract
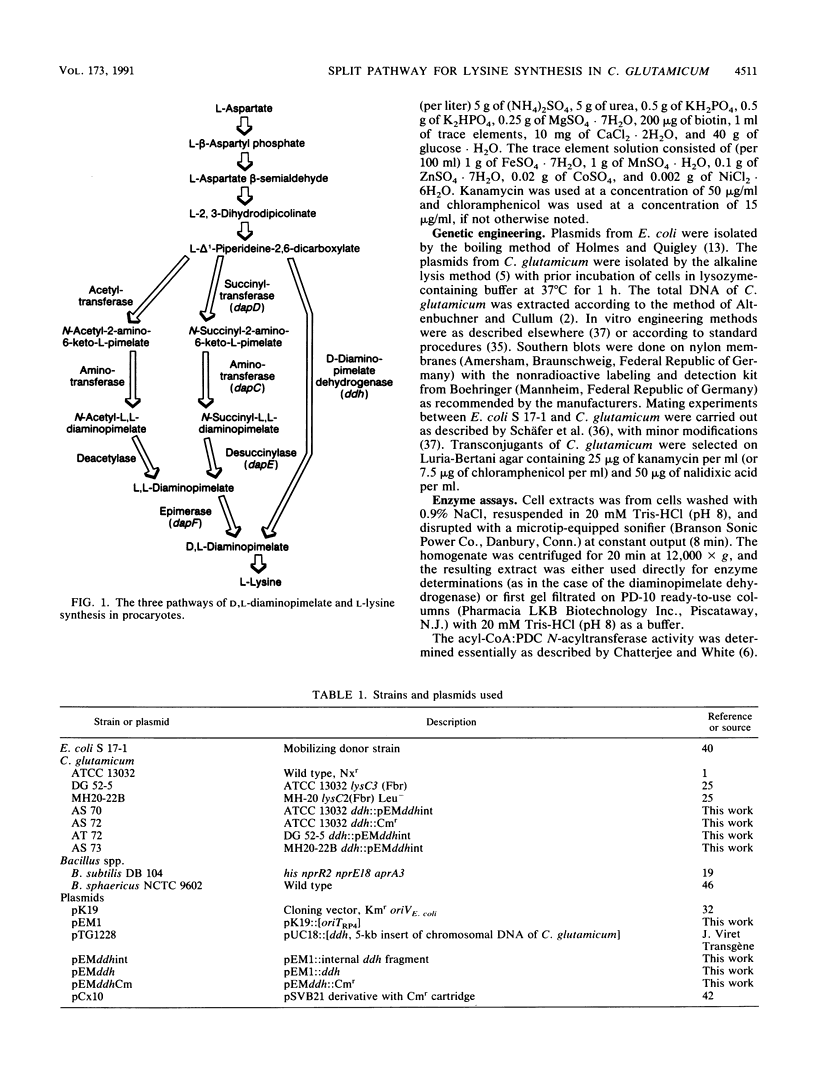
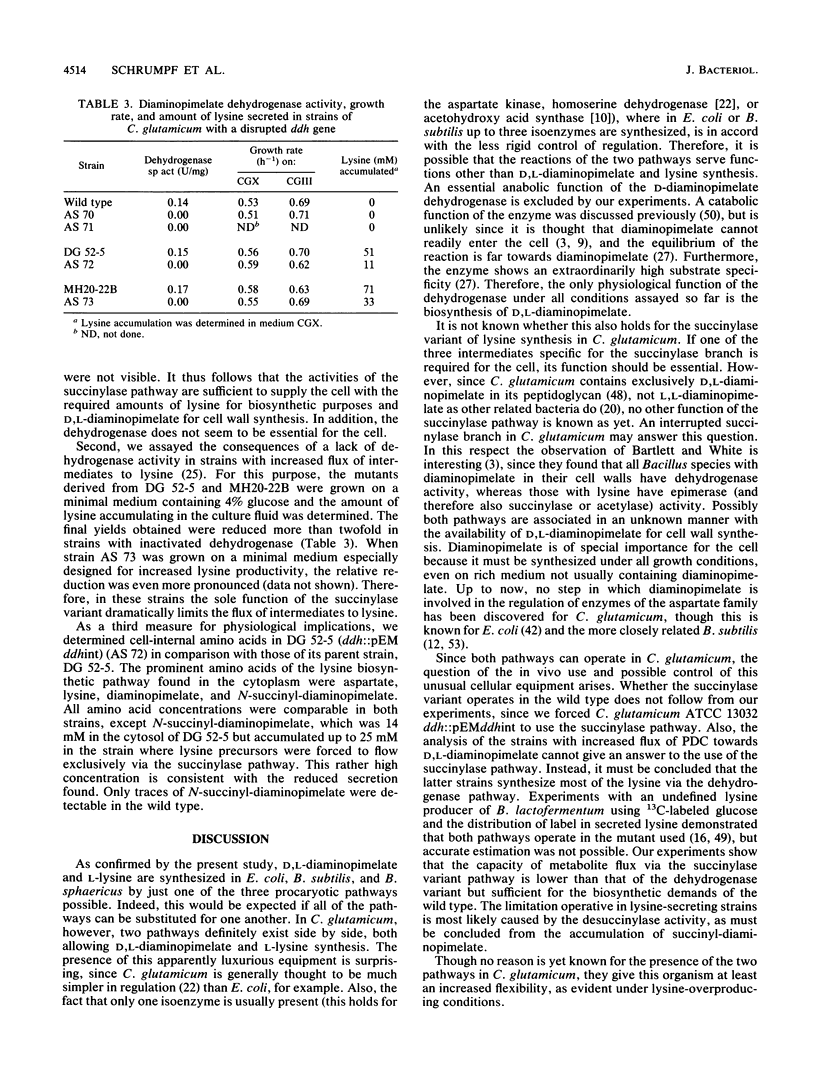
Three different pathways of D,L-diaminopimelate and L-lysine synthesis are known in procaryotes. Determinations of the corresponding enzyme activities in Escherichia coli, Bacillus subtilis, and Bacillus sphaericus verified the fact that in each of these bacteria only one of the possible pathways operates. However, in Corynebacterium glutamicum activities are present which allow in principle the use of the dehydrogenase variant and succinylase variant of lysine synthesis together. Applying gene-directed mutagenesis, various C. glutamicum strains were constructed with interrupted ddh gene. These mutants have an inactive dehydrogenase pathway but are still prototrophic, which is proof that the succinylase pathway of D,L-diaminopimelate synthesis can be utilized. In strains with an increased flow of precursors to D,L-diaminopimelate, however, the inactivation of the dehydrogenase pathway resulted in a reduced formation of lysine, with concomitant accumulation of N-succinyl-diaminopimelate in the cytosol up to a concentration of 25 mM. These data show (i) that both pathways can operate in C. glutamicum for D,L-diaminopimelate and L-lysine synthesis, (ii) that the dehydrogenase pathway is not essential, and (iii) that the dehydrogenase pathway is a prerequisite for handling an increased flow of metabolites to D,L-diaminopimelate.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Cullum J. DNA amplification and an unstable arginine gene in Streptomyces lividans 66. Mol Gen Genet. 1984;195(1-2):134–138. doi: 10.1007/BF00332735. [DOI] [PubMed] [Google Scholar]
- Berges D. A., DeWolf W. E., Jr, Dunn G. L., Newman D. J., Schmidt S. J., Taggart J. J., Gilvarg C. Studies on the active site of succinyl-CoA:tetrahydrodipicolinate N-succinyltransferase. Characterization using analogs of tetrahydrodipicolinate. J Biol Chem. 1986 May 15;261(14):6160–6167. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer J., Treptow C., Eggeling L., Sahm H. Regulation of enzymes of lysine biosynthesis in Corynebacterium glutamicum. J Gen Microbiol. 1988 Dec;134(12):3221–3229. doi: 10.1099/00221287-134-12-3221. [DOI] [PubMed] [Google Scholar]
- Cremer Josef, Eggeling Lothar, Sahm Hermann. Control of the Lysine Biosynthesis Sequence in Corynebacterium glutamicum as Analyzed by Overexpression of the Individual Corresponding Genes. Appl Environ Microbiol. 1991 Jun;57(6):1746–1752. doi: 10.1128/aem.57.6.1746-1752.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILVARG C. N-Succinyl-L-diaminopimelic acid. J Biol Chem. 1959 Nov;234:2955–2959. [PubMed] [Google Scholar]
- Graves L. M., Switzer R. L. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J Bacteriol. 1990 Jan;172(1):218–223. doi: 10.1128/jb.172.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ishino S., Mizukami T., Yamaguchi K., Katsumata R., Araki K. Nucleotide sequence of the meso-diaminopimelate D-dehydrogenase gene from Corynebacterium glutamicum. Nucleic Acids Res. 1987 May 11;15(9):3917–3917. doi: 10.1093/nar/15.9.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. N., Gilligan J. P. o-Phthaldialdehyde precolumn derivatization and reversed-phase high-performance liquid chromatography of polypeptide hydrolysates and physiological fluids. J Chromatogr. 1983 Aug 26;266:471–482. doi: 10.1016/s0021-9673(01)90918-5. [DOI] [PubMed] [Google Scholar]
- KINDLER S. H., GILVARG C. N-Succinyl-L-2,6-diaminopimelic acid deacylase. J Biol Chem. 1960 Dec;235:3532–3535. [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie R. M., Cure G. L. The cell wall composition and distribution of free mycolic acids in named strains of coryneform bacteria and in isolates from various natural sources. J Appl Bacteriol. 1977 Apr;42(2):229–252. doi: 10.1111/j.1365-2672.1977.tb00689.x. [DOI] [PubMed] [Google Scholar]
- Leadlay P. F. Free (S,S)-diaminopimelate is not an obligatory intermediate in lysine biosynthesis in Corynebacterium glutamicum. FEBS Lett. 1979 Feb 15;98(2):399–402. doi: 10.1016/0014-5793(79)80226-4. [DOI] [PubMed] [Google Scholar]
- Menkel E., Thierbach G., Eggeling L., Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989 Mar;55(3):684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misono H., Soda K. Properties of meso-alpha,epsilon-diaminopimelate D-dehydrogenase from Bacillus sphaericus. J Biol Chem. 1980 Nov 25;255(22):10599–10605. [PubMed] [Google Scholar]
- Misono H., Togawa H., Yamamoto T., Soda K. Meso-alpha,epsilon-diaminopimelate D-dehydrogenase: distribution and the reaction product. J Bacteriol. 1979 Jan;137(1):22–27. doi: 10.1128/jb.137.1.22-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERKOFSKY B., GILVARG C. N-Succinyl-L-diaminopimelic-glutamic transaminase. J Biol Chem. 1961 May;236:1432–1438. [PubMed] [Google Scholar]
- Palmieri F., Klingenberg M. Direct methods for measuring metabolite transport and distribution in mitochondria. Methods Enzymol. 1979;56:279–301. doi: 10.1016/0076-6879(79)56029-7. [DOI] [PubMed] [Google Scholar]
- Pridmore R. D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56(2-3):309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Ellman's reagent: 5,5'-dithiobis(2-nitrobenzoic acid)--a reexamination. Anal Biochem. 1979 Apr 1;94(1):75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- Rottenberg H. The measurement of membrane potential and deltapH in cells, organelles, and vesicles. Methods Enzymol. 1979;55:547–569. doi: 10.1016/0076-6879(79)55066-6. [DOI] [PubMed] [Google Scholar]
- Schwarzer A., Pühler A. Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Biotechnology (N Y) 1991 Jan;9(1):84–87. doi: 10.1038/nbt0191-84. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Simon R., Seep-Feldhaus A. H., Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990 Mar;172(3):1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms S. A., Voige W. H., Gilvarg C. Purification and characterization of succinyl-CoA: tetrahydrodipicolinate N-succinyltransferase from Escherichia coli. J Biol Chem. 1984 Mar 10;259(5):2734–2741. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sundharadas G., Gilvarg C. Biosynthesis of alpha,epsilon-diaminopimelic acid in Bacillus megaterium. J Biol Chem. 1967 Sep 10;242(17):3983–3984. [PubMed] [Google Scholar]
- Weinberger S., Gilvarg C. Bacterial distribution of the use of succinyl and acetyl blocking groups in diaminopimelic acid biosynthesis. J Bacteriol. 1970 Jan;101(1):323–324. doi: 10.1128/jb.101.1.323-324.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman J. S., Nichols J. S. Purification and properties of diaminopimelic acid epimerase from Escherichia coli. J Biol Chem. 1984 Jul 25;259(14):8907–8914. [PubMed] [Google Scholar]
- Yeh P., Sicard A. M., Sinskey A. J. General organization of the genes specifically involved in the diaminopimelate-lysine biosynthetic pathway of Corynebacterium glutamicum. Mol Gen Genet. 1988 Apr;212(1):105–111. doi: 10.1007/BF00322451. [DOI] [PubMed] [Google Scholar]
- Yeh P., Sicard A. M., Sinskey A. J. Nucleotide sequence of the lysA gene of Corynebacterium glutamicum and possible mechanisms for modulation of its expression. Mol Gen Genet. 1988 Apr;212(1):112–119. doi: 10.1007/BF00322452. [DOI] [PubMed] [Google Scholar]
- Yugari Y., Gilvarg C. The condensation step in diaminopimelate synthesis. J Biol Chem. 1965 Dec;240(12):4710–4716. [PubMed] [Google Scholar]
- Zhang J. J., Hu F. M., Chen N. Y., Paulus H. Comparison of the three aspartokinase isozymes in Bacillus subtilis Marburg and 168. J Bacteriol. 1990 Feb;172(2):701–708. doi: 10.1128/jb.172.2.701-708.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


