Abstract
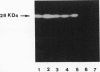
An Aeromonas hydrophila gene, named cphA, coding for a carbapenem-hydrolyzing metallo-beta-lactamase, was cloned in Escherichia coli by screening an Aeromonas genomic library for clones able to grow on imipenem-containing medium. From sequencing data, the cloned cphA gene appeared able to code for a polypeptide of 254 amino acids whose sequence includes a potential N-terminal leader sequence for targeting the protein to the periplasmic space. These data were in agreement with the molecular mass of the original Aeromonas enzyme and of the recombinant enzyme produced in E. coli, evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of crude beta-lactamase preparations followed by renaturation treatment for proteins separated in the gel and localization of protein bands showing carbapenem-hydrolyzing beta-lactamase activity by a modified iodometric technique. The deduced amino acid sequence of the CphA enzyme showed regions of partial homology with both the beta-lactamase II of Bacillus cereus and the CfiA beta-lactamase of Bacteroides fragilis. Sequence homologies were more pronounced in the regions encompassing the amino acid residues known in the enzyme of B. cereus to function as ligand-binding residues for the metal cofactor. The CphA enzyme, however, appeared to share a lower degree of similarity with the two other enzymes, which, in turn, seemed more closely related to each other. These results, therefore, suggest the existence of at least two molecular subclasses within molecular class B metallo-beta-lactamases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Bakken J. S., Sanders C. C., Clark R. B., Hori M. Beta-lactam resistance in Aeromonas spp. caused by inducible beta-lactamases active against penicillins, cephalosporins, and carbapenems. Antimicrob Agents Chemother. 1988 Sep;32(9):1314–1319. doi: 10.1128/aac.32.9.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R., Waley S. G. Cryoenzymology of Bacillus cereus beta-lactamase II. Biochemistry. 1985 Nov 19;24(24):6876–6887. doi: 10.1021/bi00345a021. [DOI] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Malamy M. H., Tally F. P. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 Nov;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fujii T., Sato K., Miyata K., Inoue M., Mitsuhashi S. Biochemical properties of beta-lactamase produced by Legionella gormanii. Antimicrob Agents Chemother. 1986 May;29(5):925–926. doi: 10.1128/aac.29.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis J. P., Sanders C. C. Purification and characterization of inducible beta-lactamases in Aeromonas spp. Antimicrob Agents Chemother. 1990 Jan;34(1):44–51. doi: 10.1128/aac.34.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen J. H., Lee J. C., Alexander G. A. Rapid penicillinase paper strip test for detection of beta-lactamase-producing Haemophilus influenzae and Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1977 Jun;11(6):1087–1088. doi: 10.1128/aac.11.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leclerc D., Asselin A. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can J Microbiol. 1989 Aug;35(8):749–753. doi: 10.1139/m89-125. [DOI] [PubMed] [Google Scholar]
- Lim H. M., Pène J. J., Shaw R. W. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 beta-lactamase II structural gene. J Bacteriol. 1988 Jun;170(6):2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C., Emanuel E. L., Gagnon J., Waley S. G. Identification of an essential glutamic acid residue in beta-lactamase II from Bacillus cereus. Biochem J. 1986 Jan 15;233(2):465–469. doi: 10.1042/bj2330465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Potvin C., Leclerc D., Tremblay G., Asselin A., Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988 Oct;214(2):241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982 Oct;22(4):564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Fujii T., Okamoto R., Inoue M., Mitsuhashi S. Biochemical properties of beta-lactamase produced by Flavobacterium odoratum. Antimicrob Agents Chemother. 1985 Apr;27(4):612–614. doi: 10.1128/aac.27.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K., King A., Phillips I. Beta-lactamases with high activity against imipenem and Sch 34343 from Aeromonas hydrophila. J Antimicrob Chemother. 1986 Jan;17(1):45–50. doi: 10.1093/jac/17.1.45. [DOI] [PubMed] [Google Scholar]
- Sutton B. J., Artymiuk P. J., Cordero-Borboa A. E., Little C., Phillips D. C., Waley S. G. An X-ray-crystallographic study of beta-lactamase II from Bacillus cereus at 0.35 nm resolution. Biochem J. 1987 Nov 15;248(1):181–188. doi: 10.1042/bj2480181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990 May;172(5):2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. J., Wu P. J., Livermore D. M. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother. 1990 May;34(5):755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]