Abstract
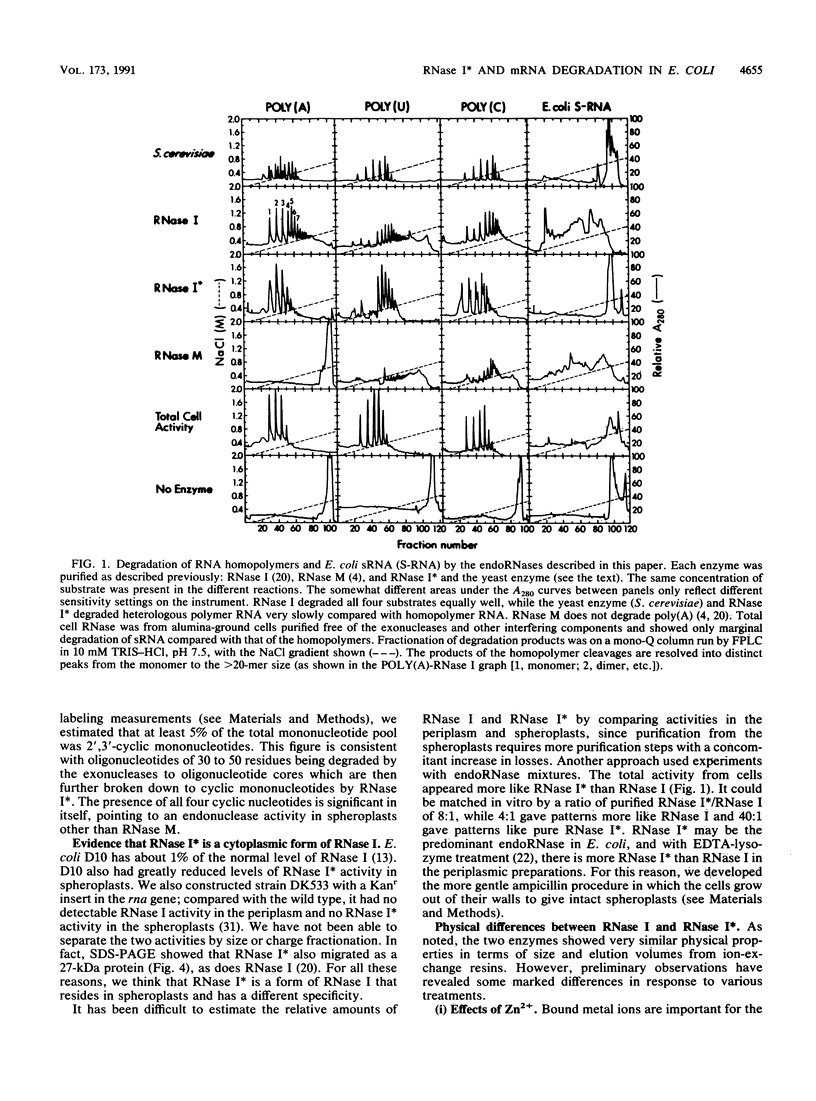
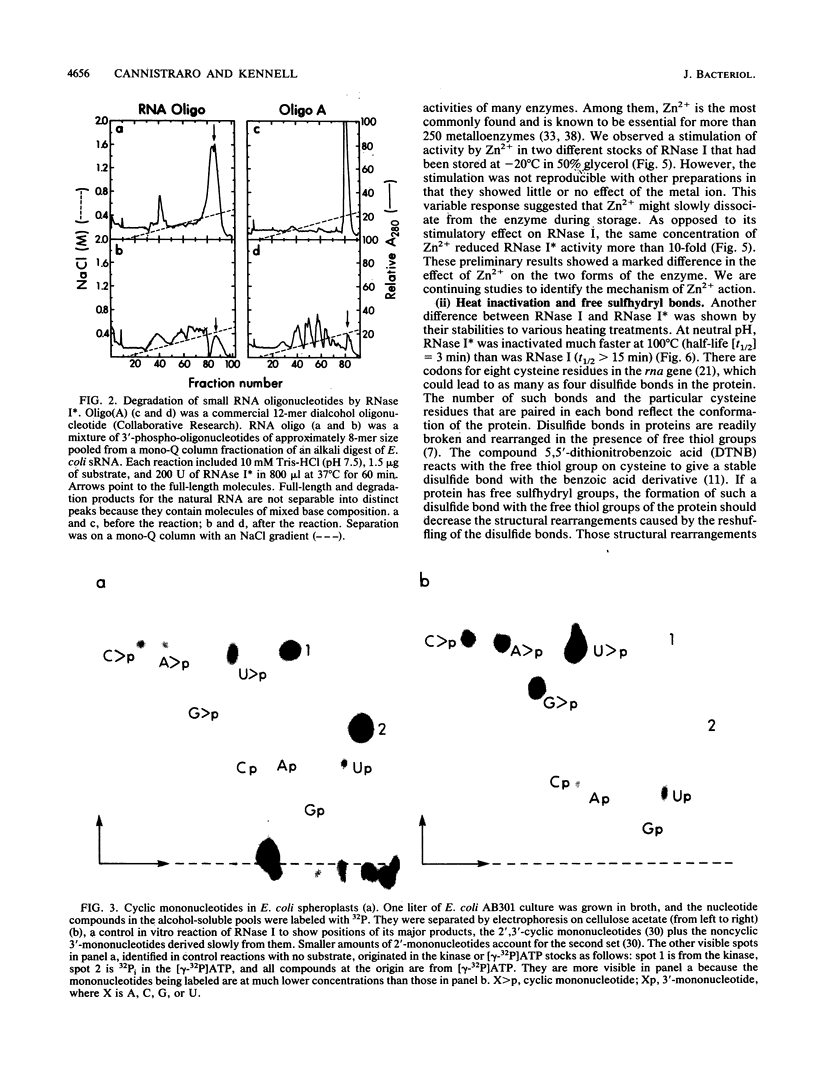
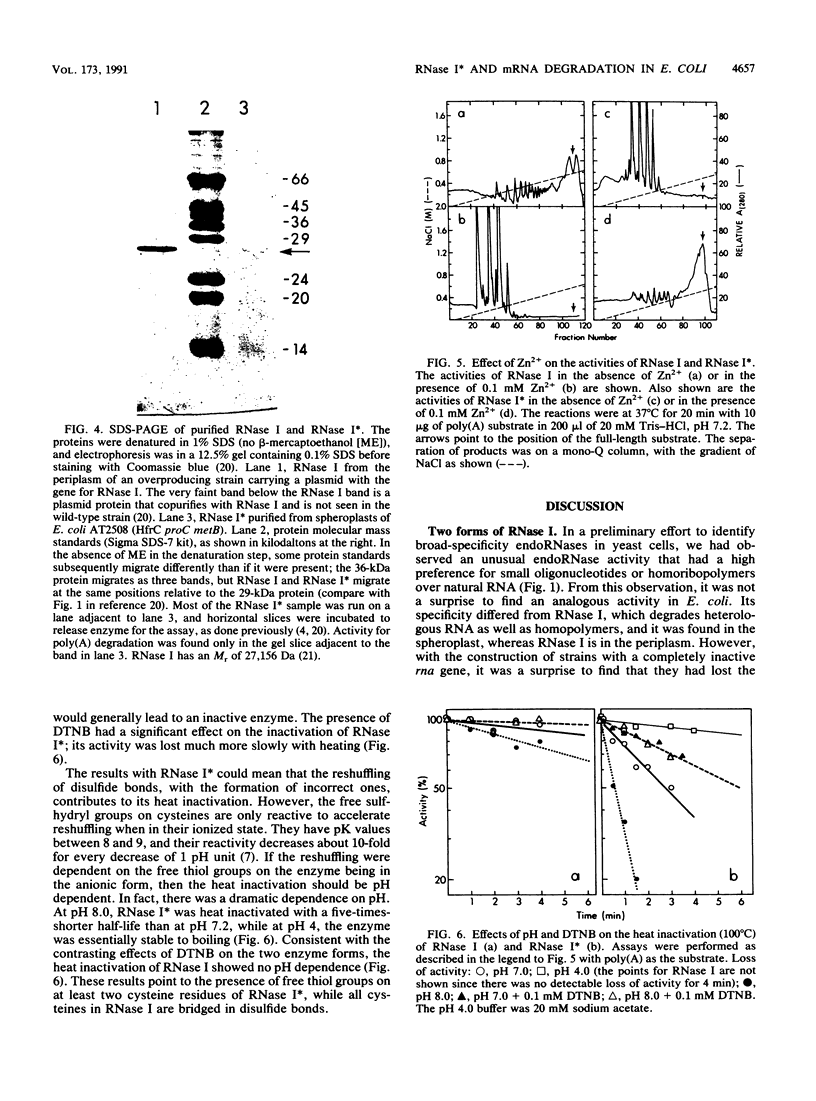
A previously unreported endoRNase present in the spheroplast fraction of Escherichia coli degraded homoribopolymers and small RNA oligonucleotides but not polymer RNA. Like the periplasmic endoRNase, RNase I, the enzyme cleaved the phosphodiester bond between any nucleotides; however, RNase I degraded polymer RNA as fast as homopolymers or oligomers. Both enzymes migrated as 27-kDa polypeptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and could not be separated by various chromatographic procedures. In rna insertion mutants, both enzymes were completely missing; the spheroplast enzyme is called RNase I*, since it must be a form of RNase I. The two forms could be distinguished by physical treatments. RNase I could be activated by Zn2+, while RNase I* was inactive in the presence of Zn2+. RNase I was inactivated very slowly at 100 degrees C over a wide pH range, while RNase I* was inactivated slowly by heat at pH 4.0 but much more rapidly as the pH was increased to 8.0. In the presence of a thiol-binding agent, the inactivation at the higher pH values was much slower. These results suggest that RNase I*, but not RNase I, has free sulfhydryl groups. RNase I* activity in the cell against a common substrate was estimated to be several times that of RNase I. All four 2',3'-phosphomonoribonucleotides were identified in the soluble pools of growing cells. Such degradative products must arise from RNase I* activity. The activity would be suited for the terminal step in mRNA degradation, the elimination of the final oligonucleotide fragments, without jeopardizing the cell RNA. An enzyme with very similar specificity was found in Saccharomyces cerevisiae, suggesting that the activity may be widespread in nature.
Full text
PDF



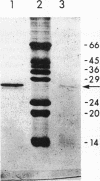
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Evidence that the 5' end of lac mRNA starts to decay as soon as it is synthesized. J Bacteriol. 1985 Feb;161(2):820–822. doi: 10.1128/jb.161.2.820-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Wice B. M., Kennell D. E. Isolating and sequencing the predominant 5'-ends of a specific mRNA in cells. II. End-labeling and sequencing. J Biochem Biophys Methods. 1985 Aug;11(2-3):163–175. doi: 10.1016/0165-022x(85)90052-1. [DOI] [PubMed] [Google Scholar]
- Datta A. K., Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975 Sep 25;250(18):7313–7319. [PubMed] [Google Scholar]
- Davis N. G., Model P. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell. 1985 Jun;41(2):607–614. doi: 10.1016/s0092-8674(85)80033-7. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- GROS F., HIATT H., GILBERT W., KURLAND C. G., RISEBROUGH R. W., WATSON J. D. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature. 1961 May 13;190:581–585. doi: 10.1038/190581a0. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Kinscherf T. G., Apirion D. Polynucleotide phosphorylase can participate in decay of mRNA in Escherichia coli in the absence of ribonuclease II. Mol Gen Genet. 1975 Sep 8;139(4):357–362. doi: 10.1007/BF00267975. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Singer M. F. The processive degradation of individual polyribonucleotide chains. II. Micrococcus lysodeikticus polynucleotide phosphorylase. J Biol Chem. 1968 Mar 10;243(5):923–927. [PubMed] [Google Scholar]
- LITTAUER U. Z., KORNBERG A. Reversible synthesis of polyribonucleotides with an enzyme from Escherichia coli. J Biol Chem. 1957 Jun;226(2):1077–1092. [PubMed] [Google Scholar]
- MacIntyre S., Freudl R., Eschbach M. L., Henning U. An artificial hydrophobic sequence functions as either an anchor or a signal sequence at only one of two positions within the Escherichia coli outer membrane protein OmpA. J Biol Chem. 1988 Dec 15;263(35):19053–19059. [PubMed] [Google Scholar]
- Meador J., 3rd, Cannon B., Cannistraro V. J., Kennell D. Purification and characterization of Escherichia coli RNase I. Comparisons with RNase M. Eur J Biochem. 1990 Feb 14;187(3):549–553. doi: 10.1111/j.1432-1033.1990.tb15336.x. [DOI] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of ribonuclease into the medium when E. coli cells are converted to spheroplasts. Biochem Biophys Res Commun. 1964;14:109–112. doi: 10.1016/0006-291x(64)90238-4. [DOI] [PubMed] [Google Scholar]
- Nossal N. G., Singer M. F. The processive degradation of individual polyribonucleotide chains. I. Escherichia coli ribonuclease II. J Biol Chem. 1968 Mar 10;243(5):913–922. [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- SEKIGUCHI M., COHEN S. S. The selective degradation of phage-induced ribonucleic acid by polynucleotide phosphorylase. J Biol Chem. 1963 Jan;238:349–356. [PubMed] [Google Scholar]
- SPAHR P. F. PURIFICATION AND PROPERTIES OF RIBONUCLEASE II FROM ESCHERICHIA COLI. J Biol Chem. 1964 Nov;239:3716–3726. [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Singer M. F., Tolbert G. Purification and properties of a potassium-activated phosphodiesterase (RNAase II) from Escherichia coli. Biochemistry. 1965 Jul;4(7):1319–1330. doi: 10.1021/bi00883a016. [DOI] [PubMed] [Google Scholar]
- Subbarao M. N., Kennell D. Evidence for endonucleolytic cleavages in decay of lacZ and lacI mRNAs. J Bacteriol. 1988 Jun;170(6):2860–2865. doi: 10.1128/jb.170.6.2860-2865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990 Jun 19;29(24):5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- WADE H. E., LOVETT S. Polynucleotide phosphorylase in ribosomes from Escherichia coli. Biochem J. 1961 Nov;81:319–328. doi: 10.1042/bj0810319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol. 1971 Sep 15;234(50):206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]