Abstract
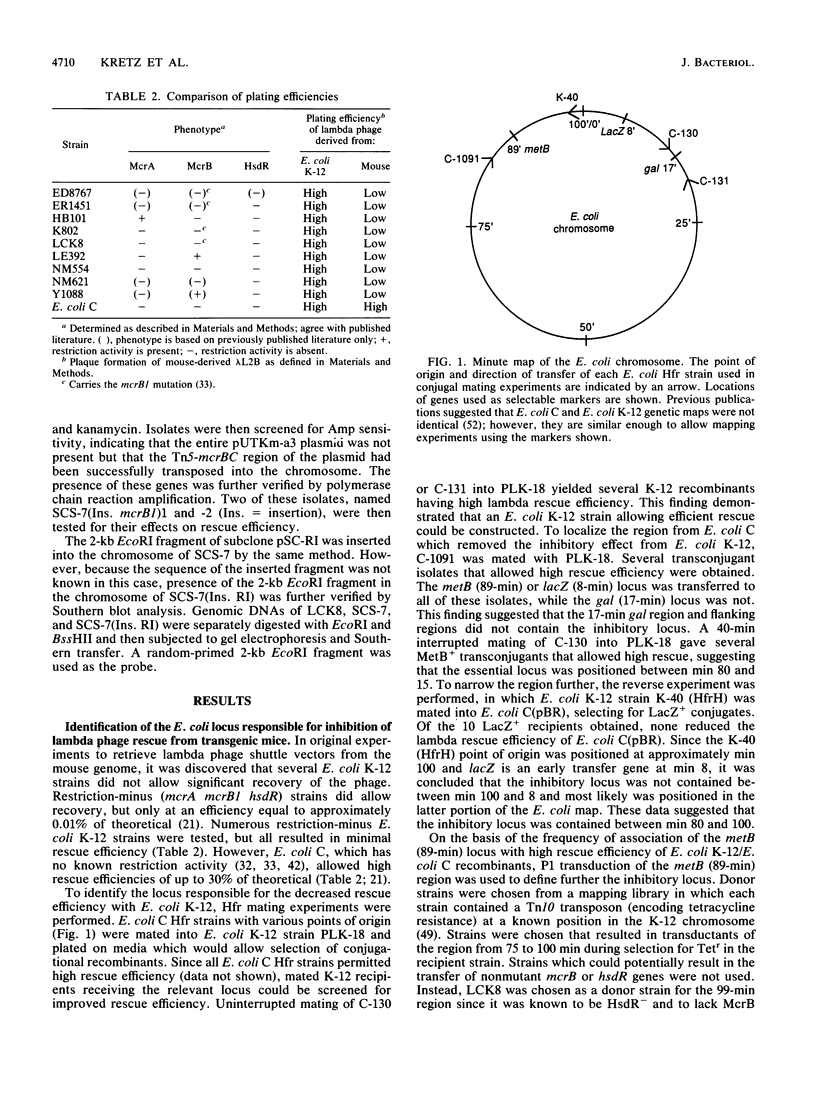
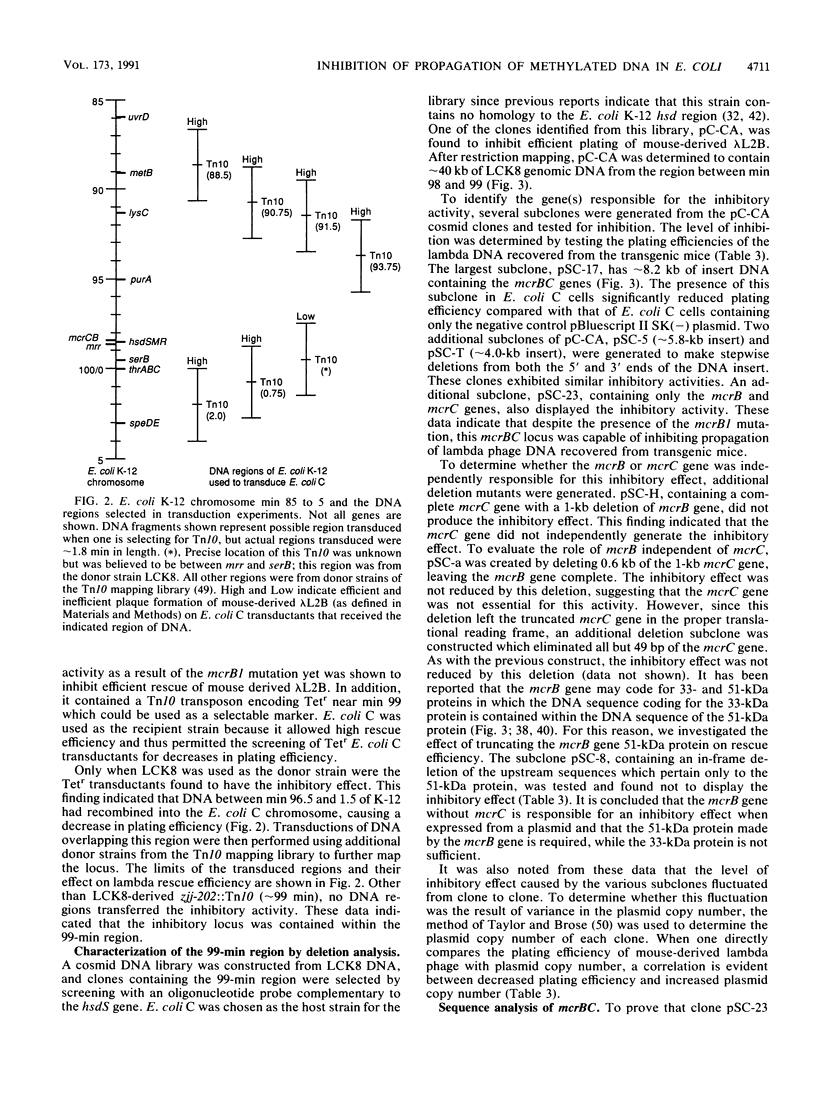
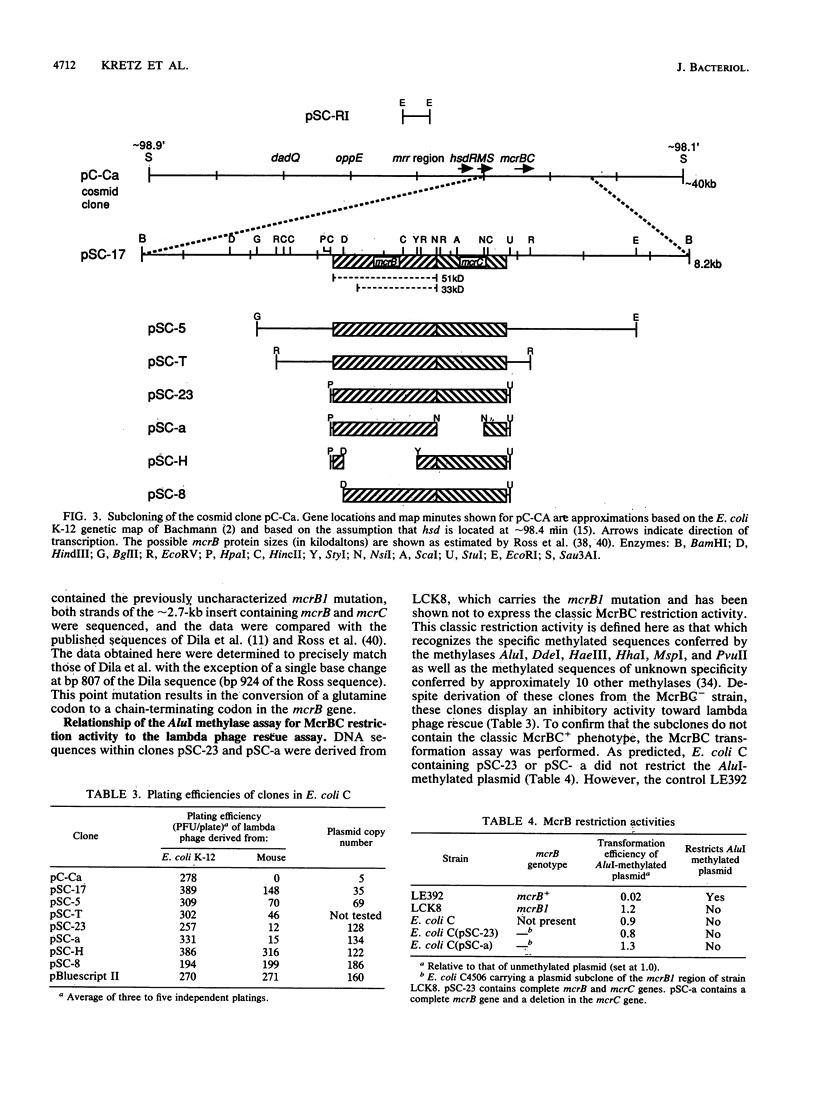
Identifying and eliminating endogenous bacterial enzyme systems can significantly increase the efficiency of propagation of eukaryotic DNA in Escherichia coli. We have recently examined one such system which inhibits the propagation of lambda DNA rescued from transgenic mouse tissues. This rescue procedure utilizes lambda packaging extracts for excision of the lambda DNA from the transgenic mouse genome, as well as E. coli cells for subsequent infection and propagation. This assay, in combination with conjugal mating, P1 transduction, and gene cloning, was used to identify and characterize the E. coli locus responsible for this difference in efficiency. It was determined that the E. coli K-12 mcrB gene when expressed on a high-copy-number plasmid can cause a decrease in rescue efficiency despite the presence of the mcrB1 mutation, which inactivates the classic McrB restriction activity. (This mutation was verified by sequence analysis.) However, this McrB1 activity is not observed when the cloned mcrB1 gene is inserted into the E. coli genome at one copy per chromosome. A second locus was identified which causes a decrease in rescue efficiency both when expressed on a high-copy-number plasmid and when inserted into the genome. The data presented here suggest that this locus is mrr and that the mrr gene product can recognize and restrict cytosine-methylated sequences. Removal of this DNA region including the mrr gene from E. coli K-12 strains allows high rescue efficiencies equal to those of E. coli C strains. These modified E. coli K-12 plating strains and lambda packaging extract strains should also allow a significant improvement in the efficiency and representation of eukaryotic genomic and cDNA libraries.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alting-Mees M. A., Short J. M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989 Nov 25;17(22):9494–9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R. M., Gregory S. A., Cooperider J. S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985 Nov;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brody H., Greener A., Hill C. W. Excision and reintegration of the Escherichia coli K-12 chromosomal element e14. J Bacteriol. 1985 Mar;161(3):1112–1117. doi: 10.1128/jb.161.3.1112-1117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther P. J., Cartwright A. L., Hocking A., Jefferson S., Ford M. D., Woodcock D. M. The effect of E. coli host strain on the consensus sequence of regions of the human L1 transposon. Nucleic Acids Res. 1989 Sep 25;17(18):7229–7239. doi: 10.1093/nar/17.18.7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dila D., Raleigh E. A. Genetic dissection of the methylcytosine-specific restriction system mcrB of Escherichia coli K-12. Gene. 1988 Dec 25;74(1):23–24. doi: 10.1016/0378-1119(88)90241-7. [DOI] [PubMed] [Google Scholar]
- Dila D., Sutherland E., Moran L., Slatko B., Raleigh E. A. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J Bacteriol. 1990 Sep;172(9):4888–4900. doi: 10.1128/jb.172.9.4888-4900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J. P., Graham M. W., Linsenmeyer M. E., Crowther P. J., Williamson M., Woodcock D. M. Effects of mcr restriction of methylated CpG islands of the L1 transposons during packaging and plating stages of mammalian genomic library construction. Gene. 1991 Feb 1;98(1):77–82. doi: 10.1016/0378-1119(91)90106-l. [DOI] [PubMed] [Google Scholar]
- Glover S. W., Colson C. Genetics of host-controlled restriction and modification in Escherichia coli. Genet Res. 1969 Apr;13(2):227–240. doi: 10.1017/s0016672300002901. [DOI] [PubMed] [Google Scholar]
- Gossen J. A., Vijg J. E. coli C: a convenient host strain for rescue of highly methylated DNA. Nucleic Acids Res. 1988 Oct 11;16(19):9343–9343. doi: 10.1093/nar/16.19.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Jessee J., Bloom F. R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990 Nov;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Rees D. J., Cheng Z., Brownlee G. G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988 Mar 25;16(6):2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S. W., Provost G. S., Kretz P. L., Dycaico M. J., Sorge J. A., Short J. M. Development of a short-term, in vivo mutagenesis assay: the effects of methylation on the recovery of a lambda phage shuttle vector from transgenic mice. Nucleic Acids Res. 1990 May 25;18(10):3007–3013. doi: 10.1093/nar/18.10.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz P. L., Reid C. H., Greener A., Short J. M. Effect of lambda packaging extract mcr restriction activity on DNA cloning. Nucleic Acids Res. 1989 Jul 11;17(13):5409–5409. doi: 10.1093/nar/17.13.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M. A novel mcrB-based Escherichia coli K-12 vector system and its use in analyzing the genetic determinants for the McrB nuclease. Gene. 1988 Dec 25;74(1):177–178. doi: 10.1016/0378-1119(88)90279-x. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Diaz R., Reiners L. Cytosine-specific DNA modification interferes with plasmid establishment in Escherichia coli K12: involvement of rglB. Mol Gen Genet. 1986 Dec;205(3):469–475. doi: 10.1007/BF00338084. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Benner J., Bloom F., Braymer H. D., DeCruz E., Dharmalingam K., Heitman J., Noyer Weidner M., Piekarowicz A., Kretz P. L. Nomenclature relating to restriction of modified DNA in Escherichia coli. J Bacteriol. 1991 Apr;173(8):2707–2709. doi: 10.1128/jb.173.8.2707-2709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A. Restriction and modification in vivo by Escherichia coli K12. Methods Enzymol. 1987;152:130–141. doi: 10.1016/0076-6879(87)52015-8. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Trimarchi R., Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989 Jun;122(2):279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R. S., Sozhamannan S., Dharmalingam K. Transposon mutagenesis and genetic mapping of the rglA and rglB loci of Escherichia coli. Mol Gen Genet. 1985;198(3):390–392. doi: 10.1007/BF00332928. [DOI] [PubMed] [Google Scholar]
- Revel H. R. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967 Apr;31(4):688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Characterization of the Escherichia coli modified cytosine restriction (mcrB) gene. Gene. 1987;61(3):277–289. doi: 10.1016/0378-1119(87)90191-0. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Identification of a second polypeptide required for McrB restriction of 5-methylcytosine-containing DNA in Escherichia coli K12. Mol Gen Genet. 1989 Apr;216(2-3):402–407. doi: 10.1007/BF00334382. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Nucleotide sequence of the McrB region of Escherichia coli K-12 and evidence for two independent translational initiation sites at the mcrB locus. J Bacteriol. 1989 Apr;171(4):1974–1981. doi: 10.1128/jb.171.4.1974-1981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross T. K., Braymer H. D. Localization of a genetic region involved in McrB restriction by Escherichia coli K-12. J Bacteriol. 1987 Apr;169(4):1757–1759. doi: 10.1128/jb.169.4.1757-1759.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E., Brose E. C. Modified Birnboim-Doly method for rapid detection of plasmid copy number. Nucleic Acids Res. 1988 Sep 26;16(18):9056–9056. doi: 10.1093/nar/16.18.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker P. A., Campbell A. J., Southern E. M., Murray N. E. Enhanced recovery and restriction mapping of DNA fragments cloned in a new lambda vector. Nucleic Acids Res. 1988 Jul 25;16(14B):6725–6736. doi: 10.1093/nar/16.14.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiman M., Bertani G., Kelly B., Sasaki I. Genetic map of Escherichia coli strain C. Mol Gen Genet. 1970;107(1):1–31. doi: 10.1007/BF00433220. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Diver W. P., Graham M., Bateman C., Baker D. J., Smith S. S. RglB facilitated cloning of highly methylated eukaryotic DNA: the human L1 transposon, plant DNA, and DNA methylated in vitro with human DNA methyltransferase. Nucleic Acids Res. 1988 May 25;16(10):4465–4482. doi: 10.1093/nar/16.10.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Doherty J., Jefferson S., DeCruz E., Noyer-Weidner M., Smith S. S., Michael M. Z., Graham M. W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989 May 11;17(9):3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



