Abstract
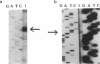
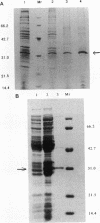
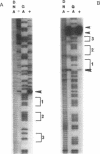
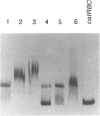
CatR, a LysR family protein, positively regulates the Pseudomonas putida catBC operon, which is required for growth on benzoate as a sole carbon source. Transcriptional studies show that the catR and catBC promoters are divergent and overlapping by 2 bp. A beta-galactosidase promoter probe vector was constructed to analyze expression from the catR and catBC promoters under induced and uninduced conditions. As predicted, the catBC promoter is expressed only under induced conditions, while the catR promoter is constitutive. CatR has been shown to specifically bind the catRBC promoter region, and this property was used to devise a purification protocol for CatR. Linear M13 DNA containing the catRBC control region was covalently bound to cyanogen bromide-activated Sepharose in order to construct a DNA affinity column. Crude extracts containing hyperproduced CatR protein were then incubated with the affinity resin under binding conditions, and the CatR protein was eluted with 1 M NaCl. CatR was also purified by heparin-agarose chromatography. This highly purified protein was used for gel retardation and hydroxyl-radical footprinting studies. From this analysis, it was shown that CatR binds upstream of the catBC promoter within the transcribed region of catR.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich T. L., Chakrabarty A. M. Transcriptional regulation, nucleotide sequence, and localization of the promoter of the catBC operon in Pseudomonas putida. J Bacteriol. 1988 Mar;170(3):1297–1304. doi: 10.1128/jb.170.3.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich T. L., Frantz B., Gill J. F., Kilbane J. J., Chakrabarty A. M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52(2-3):185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- Aldrich T. L., Rothmel R. K., Chakrabarty A. M. Identification of nucleotides critical for activity of the Pseudomonas putida catBC promoter. Mol Gen Genet. 1989 Aug;218(2):266–271. doi: 10.1007/BF00331277. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Beck C. F., Warren R. A. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988 Sep;52(3):318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X. Y., Maxon M. E., Redfield B., Glass R., Brot N., Weissbach H. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4407–4411. doi: 10.1073/pnas.86.12.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Crawford I. P. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 1990 Feb 25;18(4):979–988. doi: 10.1093/nar/18.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Genetic rearrangements in plasmids specifying total degradation of chlorinated benzoic acids. Mol Gen Genet. 1982;188(2):279–285. doi: 10.1007/BF00332688. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha M. A., Kropinski A. M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990 Jun;172(6):3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., You I. S., Chatterjee D. K., Chakrabarty A. M. Microbial degradation of halogenated compounds. Science. 1985 Apr 12;228(4696):135–142. doi: 10.1126/science.228.4696.135. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hong G. F., Burn J. E., Johnston A. W. Evidence that DNA involved in the expression of nodulation (nod) genes in Rhizobium binds to the product of the regulatory gene nodD. Nucleic Acids Res. 1987 Dec 10;15(23):9677–9690. doi: 10.1093/nar/15.23.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Lindberg F., Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J Bacteriol. 1989 Jul;171(7):3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxon M. E., Redfield B., Cai X. Y., Shoeman R., Fujita K., Fisher W., Stauffer G., Weissbach H., Brot N. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):85–89. doi: 10.1073/pnas.86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Parke D. Evolution of catabolic pathways. Biochem Soc Trans. 1976;4(3):468–472. doi: 10.1042/bst0040468. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plamann L. S., Stauffer G. V. Nucleotide sequence of the Salmonella typhimurium metR gene and the metR-metE control region. J Bacteriol. 1987 Sep;169(9):3932–3937. doi: 10.1128/jb.169.9.3932-3937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowsky P. M., Close T. J., Chimera J. A., Shaw J. J., Kado C. I. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987 Nov;169(11):5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmel R. K., Aldrich T. L., Houghton J. E., Coco W. M., Ornston L. N., Chakrabarty A. M. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J Bacteriol. 1990 Feb;172(2):922–931. doi: 10.1128/jb.172.2.922-931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Poser E. F. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J Bacteriol. 1989 Feb;171(2):837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Sukordhaman M. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J Bacteriol. 1989 Apr;171(4):1952–1959. doi: 10.1128/jb.171.4.1952-1959.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. F., Bugianesi R. L., Shen T. Y. Preparation of sepharose-bound poly (rI:rC). Biochem Biophys Res Commun. 1971 Oct 1;45(1):184–189. doi: 10.1016/0006-291x(71)90067-2. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986 Feb 15;261(5):2441–2450. [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- Wheelis M. L., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972 Feb;109(2):790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelis M. L., Stanier R. Y. The genetic control of dissimilatory pathways in Pseudomonas putida. Genetics. 1970 Oct;66(2):245–266. doi: 10.1093/genetics/66.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H., Ornston M. K., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: two-point crosses with a regulatory mutant strain. J Bacteriol. 1972 Feb;109(2):796–802. doi: 10.1128/jb.109.2.796-802.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- You I. S., Ghosal D., Gunsalus I. C. Nucleotide sequence of plasmid NAH7 gene nahR and DNA binding of the nahR product. J Bacteriol. 1988 Dec;170(12):5409–5415. doi: 10.1128/jb.170.12.5409-5415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]