Abstract
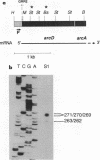
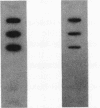
The arcDABC operon of Pseudomonas aeruginosa encodes the enzymes of the arginine deiminase pathway, which is inducible under conditions of oxygen limitation and serves to generate ATP from arginine. The 5' end of arc mRNA extracted from anaerobically grown cells was determined by S1 and primer extension mapping. The transcription initiation site was located upstream of the arcD gene and 41.5 bp downstream of the center of the sequence TTGAC....ATCAG. This sequence, termed the ANR box, is similar to the consensus FNR recognition site of Escherichia coli. Transcription of the arc operon in P. aeruginosa was strongly decreased by a deletion of the TTGAC half site or by a mutation in the anr gene, which is known to code for the FNR-like regulatory protein ANR. During a transition from aerobic to anaerobic growth conditions, the concentrations of arc mRNAs and the levels of the ArcD and ArcA proteins rose in a parallel fashion. Mutational analysis of the arc promoter region led to the conclusion that the distance between the ANR box and the -10 promoter region is important for promoter strength, whereas the -35 region does not appear to be critical for arc promoter function. These findings and previous results indicate that anaerobic induction of the arc operon occurs at the level of transcription and requires the ANR box in cis and the ANR protein in trans.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Baur H., Luethi E., Stalon V., Mercenier A., Haas D. Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of Pseudomonas aeruginosa. Eur J Biochem. 1989 Jan 15;179(1):53–60. doi: 10.1111/j.1432-1033.1989.tb14520.x. [DOI] [PubMed] [Google Scholar]
- Baur H., Stalon V., Falmagne P., Luethi E., Haas D. Primary and quaternary structure of the catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa. Extensive sequence homology with the anabolic ornithine carbamoyltransferases of Escherichia coli. Eur J Biochem. 1987 Jul 1;166(1):111–117. doi: 10.1111/j.1432-1033.1987.tb13489.x. [DOI] [PubMed] [Google Scholar]
- Bell A. I., Cole J. A., Busby S. J. Molecular genetic analysis of an FNR-dependent anaerobically inducible Escherichia coli promoter. Mol Microbiol. 1990 Oct;4(10):1753–1763. doi: 10.1111/j.1365-2958.1990.tb00553.x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Schilling-Cordaro C., Mergia A., Houck C. M. A new technique for genetic engineering of Agrobacterium Ti plasmid. Plasmid. 1983 Jul;10(1):21–30. doi: 10.1016/0147-619x(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Davies K. J., Lloyd D., Boddy L. The effect of oxygen on denitrification in Paracoccus denitrificans and Pseudomonas aeruginosa. J Gen Microbiol. 1989 Sep;135(9):2445–2451. doi: 10.1099/00221287-135-9-2445. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiglmeier K., Honoré N., Iuchi S., Lin E. C., Cole S. T. Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol. 1989 Jul;3(7):869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh R., Watson J. M., Haas D. Construction of recombination-deficient strains of Pseudomonas aeruginosa. Mol Gen Genet. 1983;191(2):334–337. doi: 10.1007/BF00334835. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991 Mar;173(5):1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Green J., Trageser M., Six S., Unden G., Guest J. R. Characterization of the FNR protein of Escherichia coli, an iron-binding transcriptional regulator. Proc Biol Sci. 1991 May 22;244(1310):137–144. doi: 10.1098/rspb.1991.0062. [DOI] [PubMed] [Google Scholar]
- Haas D., Watson J., Krieg R., Leisinger T. Isolation of an Hfr donor of Pseudomonas aeruginosa PAO by insertion of the plasmid RP1 into the tryptophan synthase gene. Mol Gen Genet. 1981;182(2):240–244. doi: 10.1007/BF00269664. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke H., Günther I., Binder F. A novel cloning vector for the direct selection of recombinant DNA in E. coli. Gene. 1982 Sep;19(2):231–234. doi: 10.1016/0378-1119(82)90011-7. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Kiley P. J., Reznikoff W. S. Fnr mutants that activate gene expression in the presence of oxygen. J Bacteriol. 1991 Jan;173(1):16–22. doi: 10.1128/jb.173.1.16-22.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Baur H., Gamper M., Brunner F., Villeval D., Mercenier A., Haas D. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene. 1990 Mar 1;87(1):37–43. doi: 10.1016/0378-1119(90)90493-b. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Mercenier A., Haas D. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J Gen Microbiol. 1986 Oct;132(10):2667–2675. doi: 10.1099/00221287-132-10-2667. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville S. B., Gunsalus R. P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990 Nov 5;265(31):18733–18736. [PubMed] [Google Scholar]
- Mercenier A., Simon J. P., Vander Wauven C., Haas D., Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980 Oct;144(1):159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Minton N. P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984 Nov;31(1-3):269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- Morgan A. F. Transduction of Pseudomonas aeruginosa with a mutant of bacteriophage E79. J Bacteriol. 1979 Jul;139(1):137–140. doi: 10.1128/jb.139.1.137-140.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Reimmann C., Rella M., Haas D. Integration of replication-defective R68.45-like plasmids into the Pseudomonas aeruginosa chromosome. J Gen Microbiol. 1988 Jun;134(6):1515–1523. doi: 10.1099/00221287-134-6-1515. [DOI] [PubMed] [Google Scholar]
- Rella M., Mercenier A., Haas D. Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene. 1985;33(3):293–303. doi: 10.1016/0378-1119(85)90237-9. [DOI] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Gaston K. L., Bell A. I., Roberts R. E., Busby S. J., Guest J. R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990 Nov;4(11):1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Roberts R. E., Guest J. R. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR-regulated gene expression. Mol Microbiol. 1989 May;3(5):601–608. doi: 10.1111/j.1365-2958.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Holloway B. W. A mutant sex factor of Pseudomonas aeruginosa. Genet Res. 1972 Feb;19(1):91–108. doi: 10.1017/s0016672300014294. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubben D., Schmitt R. Tn1721 derivatives for transposon mutagenesis, restriction mapping and nucleotide sequence analysis. Gene. 1986;41(2-3):145–152. doi: 10.1016/0378-1119(86)90093-4. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Holst B., Søgaard-Andersen L., Martinussen J., Nesvera J., Douthwaite S. R. Design of cAMP-CRP-activated promoters in Escherichia coli. Mol Microbiol. 1991 Feb;5(2):433–437. doi: 10.1111/j.1365-2958.1991.tb02126.x. [DOI] [PubMed] [Google Scholar]
- Van Hartingsveldt J., Marinus M. G., Stouthamer A. H. Mutants of Pseudomonas aeruginosa bblocked in nitrate or nitrite dissimilation. Genetics. 1971 Apr;67(4):469–482. doi: 10.1093/genetics/67.4.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C., Piérard A., Kley-Raymann M., Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984 Dec;160(3):928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtli M., Hütter R. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol Gen Genet. 1987 Jun;208(1-2):195–203. doi: 10.1007/BF00330442. [DOI] [PubMed] [Google Scholar]
- Walker M. S., DeMoss J. A. Promoter sequence requirements for Fnr-dependent activation of transcription of the narGHJI operon. Mol Microbiol. 1991 Feb;5(2):353–360. doi: 10.1111/j.1365-2958.1991.tb02116.x. [DOI] [PubMed] [Google Scholar]