Abstract
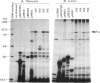
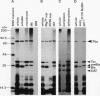
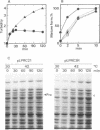
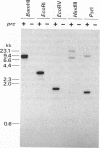
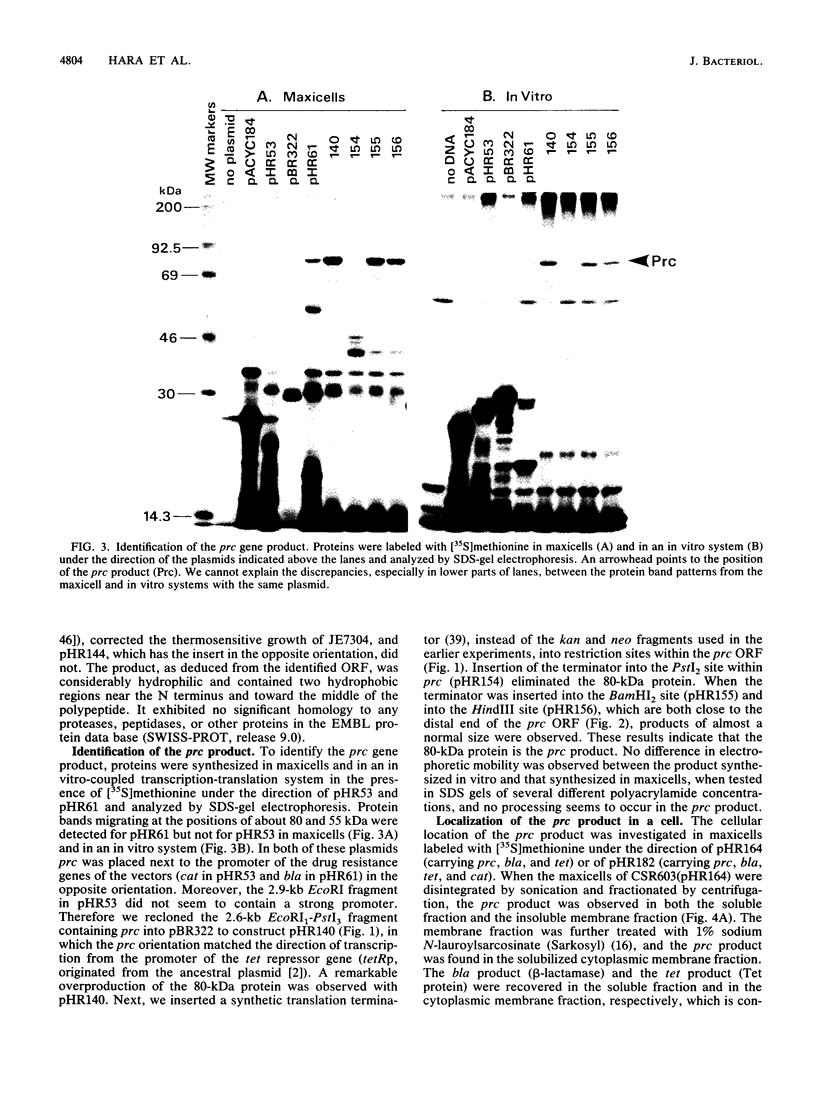
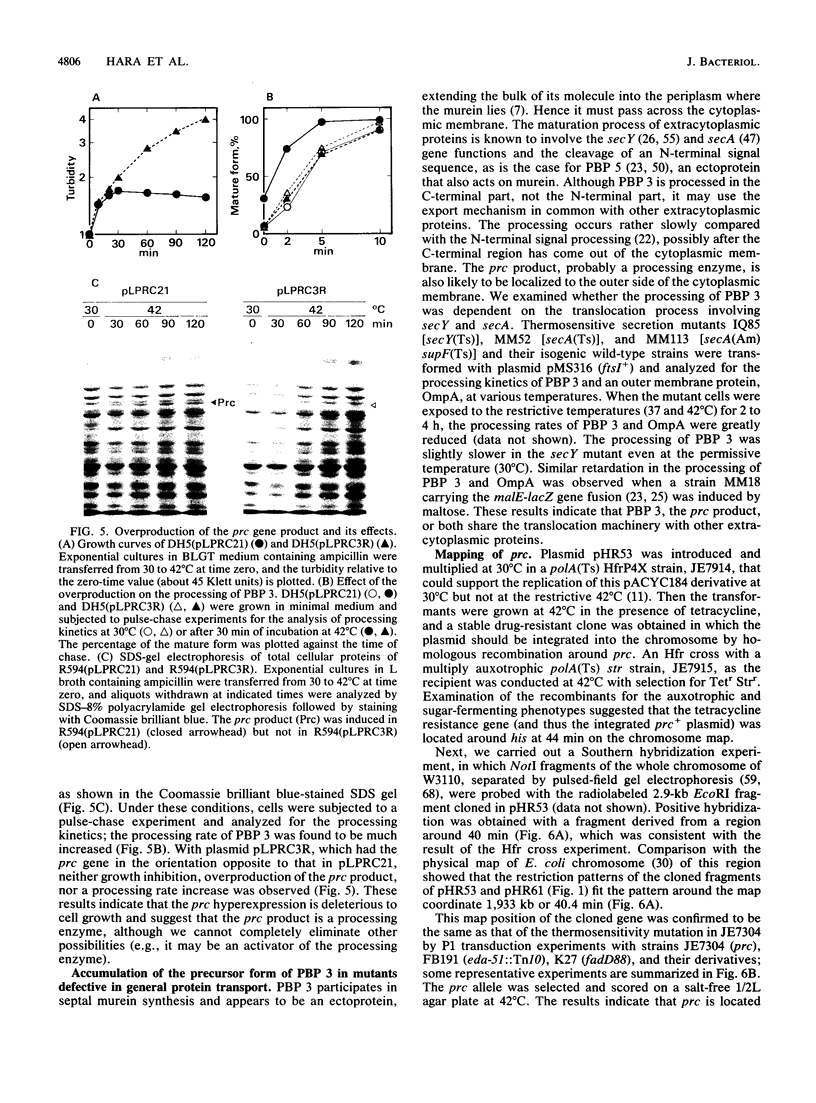
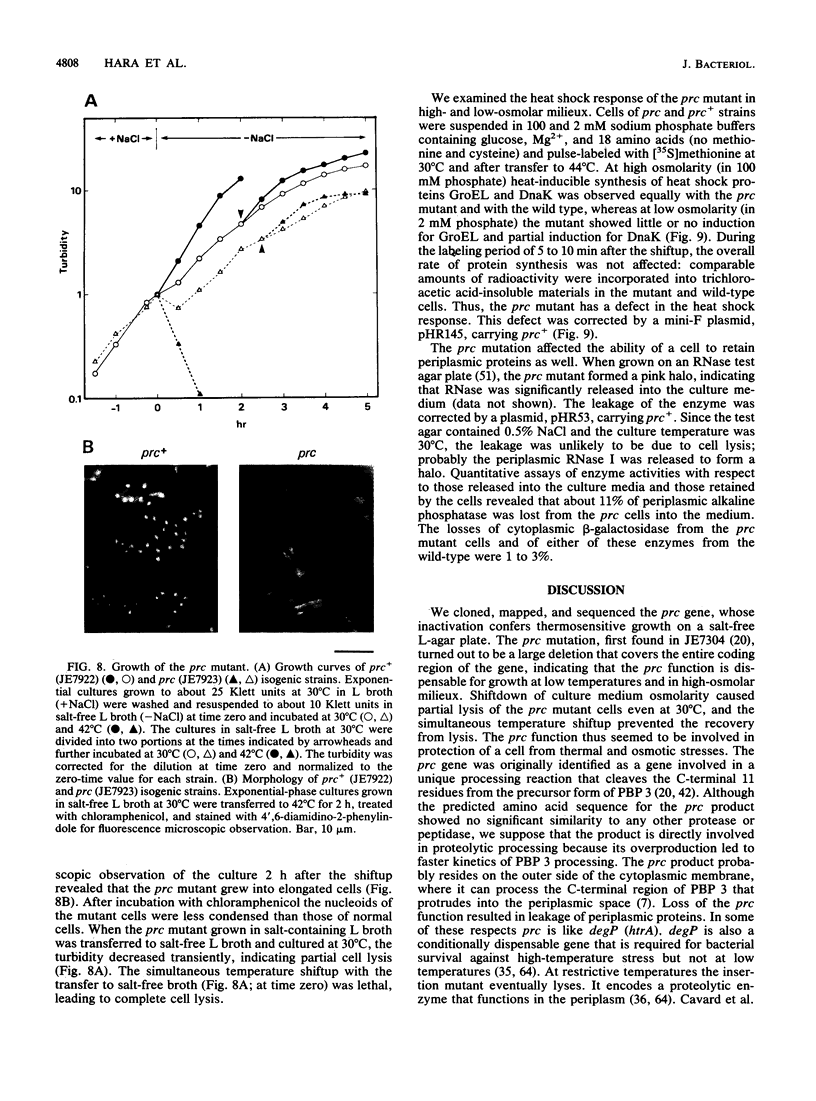
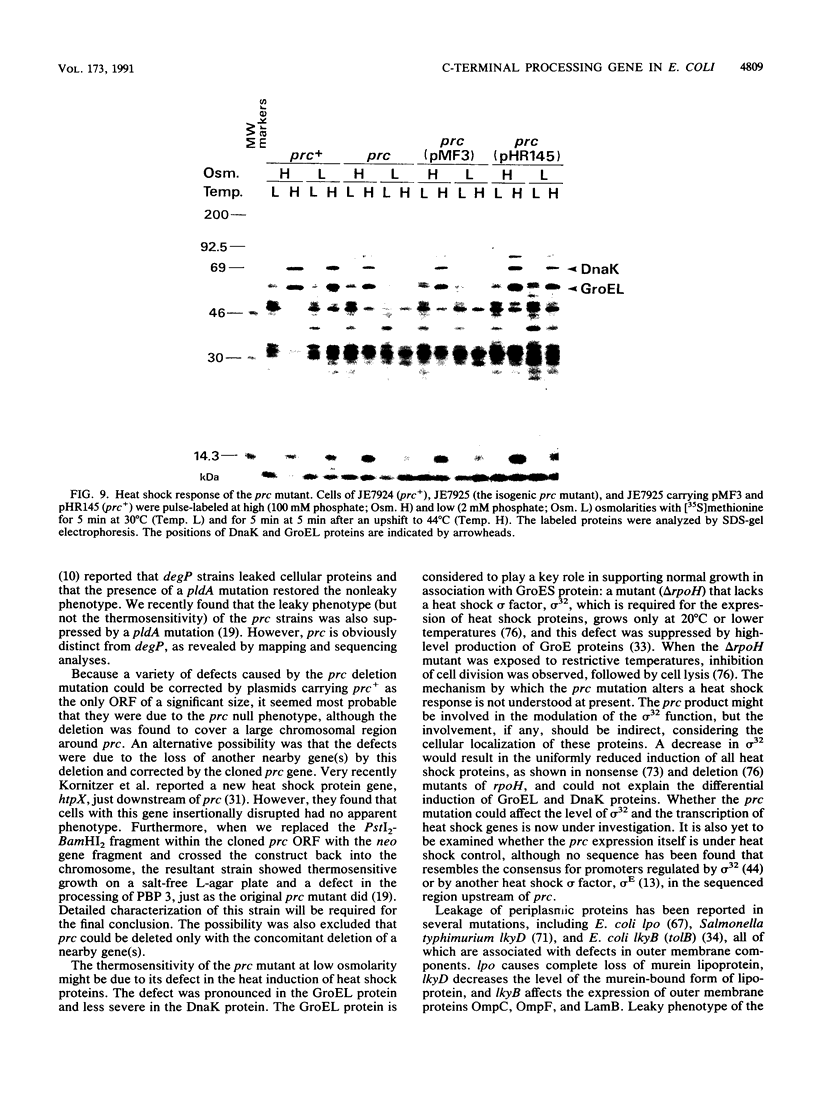
The prc gene, which is involved in cleavage of the C-terminal peptide from the precursor form of penicillin-binding protein 3 (PBP 3) of Escherichia coli, was cloned and mapped at 40.4 min on the chromosome. The gene product was identified as a protein of about 80 kDa in maxicell and in vitro systems. Fractionation of the maxicells producing the product suggested that the product was associated with the periplasmic side of the cytoplasmic membrane. This was consistent with the notion that the C-terminal processing of PBP 3 probably occurs outside the cytoplasmic membrane: the processing was found to be dependent on the secY and secA functions, indicating that the prc product or PBP 3 or both share the translocation machinery with other extracytoplasmic proteins. DNA sequencing analysis of the prc gene region identified an open reading frame, with two possible translational starts 6 bp apart from each other, that could code for a product with a calculated molecular weight of 76,667 or 76,432. The prc mutant was sensitive to thermal and osmotic stresses. Southern analysis of the chromosomal DNA of the mutant unexpectedly revealed that the mutation was a deletion of the entire prc gene and thus that the prc gene is conditionally dispensable. The mutation resulted in greatly reduced heat shock response at low osmolarity and in leakage of periplasmic proteins.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbás P., Soberón X., Merino E., Zurita M., Lomeli H., Valle F., Flores N., Bolivar F. Plasmid vector pBR322 and its special-purpose derivatives--a review. Gene. 1986;50(1-3):3–40. doi: 10.1016/0378-1119(86)90307-0. [DOI] [PubMed] [Google Scholar]
- Bassford P. J., Jr, Silhavy T. J., Beckwith J. R. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J Bacteriol. 1979 Jul;139(1):19–31. doi: 10.1128/jb.139.1.19-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belder J. B., Nguyen-Distèche M., Houba-Herin N., Ghuysen J. M., Maruyama I. N., Hara H., Hirota Y., Inouye M. Overexpression, solubilization and refolding of a genetically engineered derivative of the penicillin-binding protein 3 of Escherichia coli K12. Mol Microbiol. 1988 Jul;2(4):519–525. doi: 10.1111/j.1365-2958.1988.tb00058.x. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Model P. A prokaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5200–5204. doi: 10.1073/pnas.79.17.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bowler L. D., Spratt B. G. Membrane topology of penicillin-binding protein 3 of Escherichia coli. Mol Microbiol. 1989 Sep;3(9):1277–1286. doi: 10.1111/j.1365-2958.1989.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Carson M. J., Barondess J., Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991 Apr;173(7):2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J., Arber W. E. coli K-12 pel mutants, which block phage lambda DNA injection, coincide with ptsM, which determines a component of a sugar transport system. Mol Gen Genet. 1978 Apr 25;161(1):1–8. doi: 10.1007/BF00266608. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Gross C. A. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989 Sep;3(9):1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- Erni B., Zanolari B., Kocher H. P. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J Biol Chem. 1987 Apr 15;262(11):5238–5247. [PubMed] [Google Scholar]
- Erni B., Zanolari B. The mannose-permease of the bacterial phosphotransferase system. Gene cloning and purification of the enzyme IIMan/IIIMan complex of Escherichia coli. J Biol Chem. 1985 Dec 15;260(29):15495–15503. [PubMed] [Google Scholar]
- Filip C., Fletcher G., Wulff J. L., Earhart C. F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973 Sep;115(3):717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Nishimura A., Serizawa H. Genetic mapping of the Escherichia coli gene for the stringent starvation protein and its dispensability for normal cell growth. Mol Gen Genet. 1988 Mar;211(3):515–519. doi: 10.1007/BF00425709. [DOI] [PubMed] [Google Scholar]
- Hara H., Nishimura Y., Kato J., Suzuki H., Nagasawa H., Suzuki A., Hirota Y. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989 Nov;171(11):5882–5889. doi: 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Hara H., Suzuki H., Hirota Y. Lipid modification of Escherichia coli penicillin-binding protein 3. J Bacteriol. 1988 Nov;170(11):5392–5395. doi: 10.1128/jb.170.11.5392-5395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., Jackson M., Bassford P. J., Sinden D., Holland I. B. Insertion of a MalE beta-galactosidase fusion protein into the envelope of Escherichia coli disrupts biogenesis of outer membrane proteins and processing of inner membrane proteins. J Bacteriol. 1982 Oct;152(1):133–139. doi: 10.1128/jb.152.1.133-139.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houba-Hérin N., Hara H., Inouye M., Hirota Y. Binding of penicillin to thiol-penicillin-binding protein 3 of Escherichia coli: identification of its active site. Mol Gen Genet. 1985;201(3):499–504. doi: 10.1007/BF00331346. [DOI] [PubMed] [Google Scholar]
- Ito K., Bassford P. J., Jr, Beckwith J. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell. 1981 Jun;24(3):707–717. doi: 10.1016/0092-8674(81)90097-0. [DOI] [PubMed] [Google Scholar]
- Ito K., Wittekind M., Nomura M., Shiba K., Yura T., Miura A., Nashimoto H. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell. 1983 Mar;32(3):789–797. doi: 10.1016/0092-8674(83)90065-x. [DOI] [PubMed] [Google Scholar]
- Jones C., Holland I. B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Suzuki H. Escherichia coli parA is an allele of the gyrB gene. Mol Gen Genet. 1989 May;217(1):178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kornitzer D., Teff D., Altuvia S., Oppenheim A. B. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J Bacteriol. 1991 May;173(9):2944–2953. doi: 10.1128/jb.173.9.2944-2953.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A., Oliver D. B., Beckwith J. Signal sequence mutations disrupt feedback between secretion of an exported protein and its synthesis in E. coli. 1984 Apr 26-May 2Nature. 308(5962):863–864. doi: 10.1038/308863a0. [DOI] [PubMed] [Google Scholar]
- Kusukawa N., Yura T. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 1988 Jul;2(7):874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- Lazzaroni J. C., Fognini-Lefebvre N., Portalier R. C. Cloning of the lkyB (tolB) gene of Escherichia coli K12 and characterization of its product. Mol Gen Genet. 1986 Aug;204(2):285–288. doi: 10.1007/BF00425511. [DOI] [PubMed] [Google Scholar]
- Lipinska B., Fayet O., Baird L., Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989 Mar;171(3):1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska B., Zylicz M., Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990 Apr;172(4):1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J. J., Kline B. C. Restriction endonuclease mapping and mutagenesis of the F sex factor replication region. Mol Gen Genet. 1977 Apr 29;152(3):175–182. doi: 10.1007/BF00268815. [DOI] [PubMed] [Google Scholar]
- Maruyama I. N., Horikoshi K., Nagase Y., Soma M., Nobuhara M., Yasuda S., Hirota Y. A synthetic translation-terminator gene. A tool for dissecting the translation direction of a gene. Gene Anal Tech. 1989 May-Jun;6(3):57–61. doi: 10.1016/0735-0651(89)90011-3. [DOI] [PubMed] [Google Scholar]
- Nagasawa H., Sakagami Y., Suzuki A., Suzuki H., Hara H., Hirota Y. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989 Nov;171(11):5890–5893. doi: 10.1128/jb.171.11.5890-5893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Nucifora G., Chu L., Misra T. K., Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci U S A. 1989 May;86(10):3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981 Sep;25(3):765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- Overath P., Pauli G., Schairer H. U. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969 Feb;7(4):559–574. [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Pratt J. M., Holland I. B., Spratt B. G. Precursor forms of penicillin-binding proteins 5 and 6 of E. coli cytoplasmic membrane. Nature. 1981 Sep 24;293(5830):307–309. doi: 10.1038/293307a0. [DOI] [PubMed] [Google Scholar]
- Quaas R., Landt O., Grunert H. P., Beineke M., Hahn U. Indicator plates for rapid detection of ribonuclease T1 secreting Escherichia coli clones. Nucleic Acids Res. 1989 Apr 25;17(8):3318–3318. doi: 10.1093/nar/17.8.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Filamentous phage pre-coat is an integral membrane protein: analysis by a new method of membrane preparation. Cell. 1982 Jan;28(1):177–184. doi: 10.1016/0092-8674(82)90387-7. [DOI] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. J Mol Biol. 1981 May 5;148(1):45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Scandella D., Arber W. An Escherichia coli mutant which inhibits the injection of phage lambda DNA. Virology. 1974 Apr;58(2):504–513. doi: 10.1016/0042-6822(74)90084-1. [DOI] [PubMed] [Google Scholar]
- Shiba K., Ito K., Yura T., Cerretti D. P. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 1984 Mar;3(3):631–635. doi: 10.1002/j.1460-2075.1984.tb01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Lloyd R. G., Benson F. E., Attfield P. V. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol Gen Genet. 1984;194(1-2):322–329. doi: 10.1007/BF00383535. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977 Jul;131(1):293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storts D. R., Aparicio O. M., Schoemaker J. M., Markovitz A. Overproduction and identification of the ftsQ gene product, an essential cell division protein in Escherichia coli K-12. J Bacteriol. 1989 Aug;171(8):4290–4297. doi: 10.1128/jb.171.8.4290-4297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch K. L., Johnson K., Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989 May;171(5):2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Iino T. In vitro synthesis of phase-specific flagellin of Salmonella. J Mol Biol. 1973 Nov 25;81(1):57–70. doi: 10.1016/0022-2836(73)90247-7. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nishimura Y., Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci U S A. 1978 Feb;75(2):664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Nishimura Y., Yasuda S., Nishimura A., Yamada M., Hirota Y. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol Gen Genet. 1978 Nov 16;167(1):1–9. doi: 10.1007/BF00270315. [DOI] [PubMed] [Google Scholar]
- Tabata S., Higashitani A., Takanami M., Akiyama K., Kohara Y., Nishimura Y., Nishimura A., Yasuda S., Hirota Y. Construction of an ordered cosmid collection of the Escherichia coli K-12 W3110 chromosome. J Bacteriol. 1989 Feb;171(2):1214–1218. doi: 10.1128/jb.171.2.1214-1218.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weigand R. A., Vinci K. D., Rothfield L. I. Morphogenesis of the bacterial division septum: a new class of septation-defective mutants. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1882–1886. doi: 10.1073/pnas.73.6.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T., Yura T. Genetic control of heat-shock protein synthesis and its bearing on growth and thermal resistance in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1982 Feb;79(3):860–864. doi: 10.1073/pnas.79.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhou Y. N., Kusukawa N., Erickson J. W., Gross C. A., Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J Bacteriol. 1988 Aug;170(8):3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]