Abstract
A procedure for the purification of a very hydrophobic lipase from Pseudomonas sp. strain ATCC 21808 was elaborated by avoiding the use of long-chain detergents in view of subsequent crystallization of the enzyme. The purification procedure included chromatography on Q-Sepharose in the presence of n-octyl-beta-D-glucopyranoside, Ca2+ precipitation of fatty acids, and Octyl-Sepharose chromatography. The enzyme was purified 260-fold to a yield of 35% and a specific activity of 3,300 U/mg. The molecular weight was determined as 35,000; a polyacrylamide gel under nondenaturing conditions revealed a band at 110,000, and the isoelectric point proved to be at 4.5 to 4.6. The lipase crystallized with different salts and ethylene glycol polymers in the presence of n-octyl-beta-D-glucopyranoside and one alkyloligooxyethylene compound (CxEy) in the range from C5E2 to C8E4. The crystals diffract to a resolution of about 0.25 nm. Precession photographs revealed that they belong to space group C2 with lattice constants of a = 9.27 nm, b = 4.74 nm, c = 8.65 nm, and beta = 122.3 degrees, indicating a cell content of one molecule per asymmetric unit of the crystal. In hydrolysis of triglycerides, the lipase showed substrate specificity for saturated fatty acids from C6 to C12 and unsaturated long-chain fatty acids. Monoglycerides were hydrolyzed very slowly. The N-terminal sequence is identical to that of the lipase from Pseudomonas cepacia. Treatment with diethyl-p-nitrophenylphosphate affected the activities toward triolein and p-nitrophenylacetate to the same extent and with the same velocity.
Full text
PDF

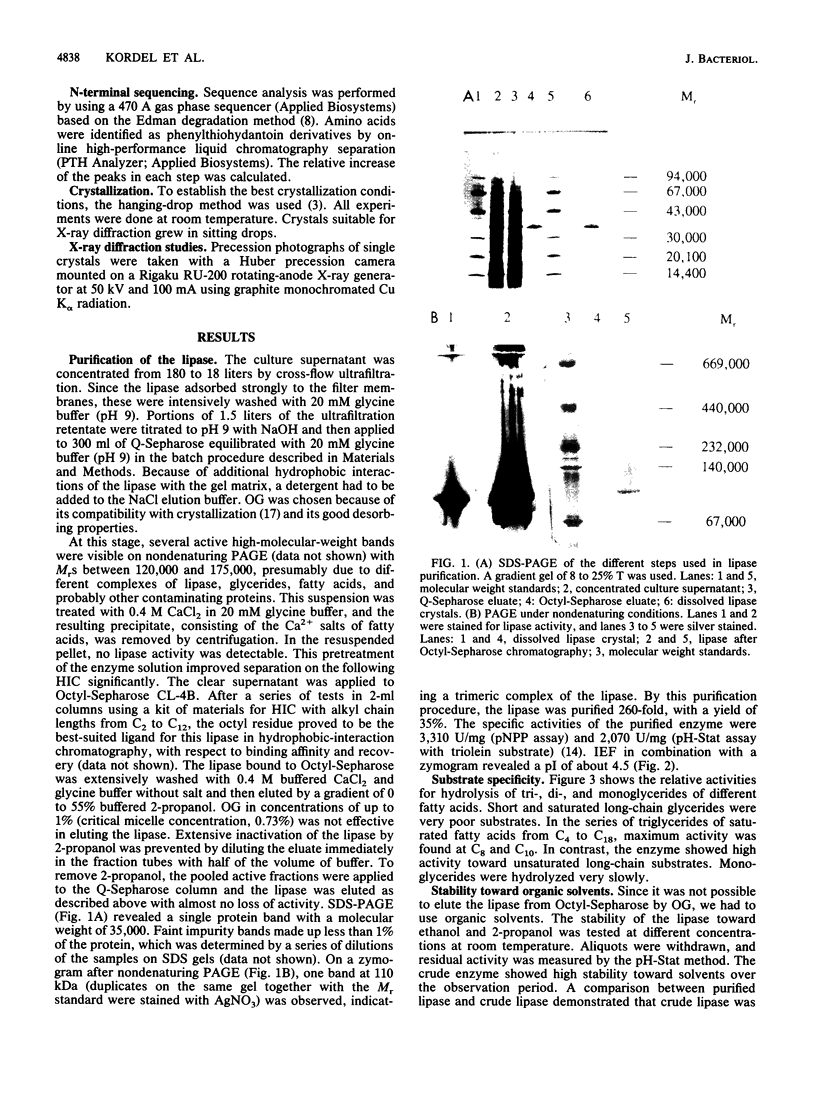
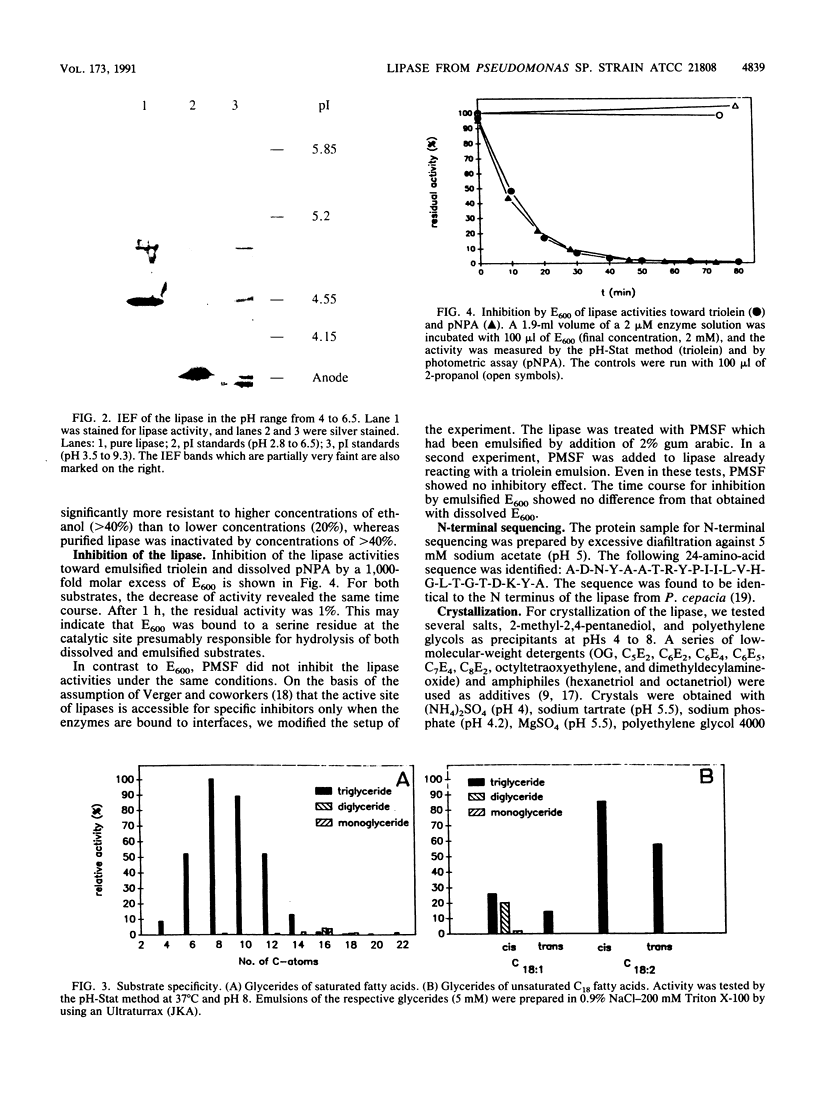


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonian E. Recent advances in the purification, characterization and structure determination of lipases. Lipids. 1988 Dec;23(12):1101–1106. doi: 10.1007/BF02535273. [DOI] [PubMed] [Google Scholar]
- Aoyama S., Yoshida N., Inouye S. Cloning, sequencing and expression of the lipase gene from Pseudomonas fragi IFO-12049 in E. coli. FEBS Lett. 1988 Dec 19;242(1):36–40. doi: 10.1016/0014-5793(88)80980-3. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brady L., Brzozowski A. M., Derewenda Z. S., Dodson E., Dodson G., Tolley S., Turkenburg J. P., Christiansen L., Huge-Jensen B., Norskov L. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature. 1990 Feb 22;343(6260):767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- Chapus C., Rovery M., Sarda L., Verger R. Minireview on pancreatic lipase and colipase. Biochimie. 1988 Sep;70(9):1223–1234. doi: 10.1016/0300-9084(88)90188-5. [DOI] [PubMed] [Google Scholar]
- Eiselé J. L., Rosenbusch J. P. Crystallization of porin using short chain phospholipids. J Mol Biol. 1989 Mar 5;206(1):209–212. doi: 10.1016/0022-2836(89)90533-0. [DOI] [PubMed] [Google Scholar]
- Furth A. J. Removing unbound detergent from hydrophobic proteins. Anal Biochem. 1980 Dec;109(2):207–215. doi: 10.1016/0003-2697(80)90638-7. [DOI] [PubMed] [Google Scholar]
- Gargouri Y., Moreau H., Verger R. Gastric lipases: biochemical and physiological studies. Biochim Biophys Acta. 1989 Dec 18;1006(3):255–271. doi: 10.1016/0005-2760(89)90012-x. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Otani Y., Hashimoto Y., Takagi Y. Molecular cloning and nucleotide sequence of the lipase gene from Pseudomonas fragi. Biochem Biophys Res Commun. 1986 Nov 26;141(1):185–190. doi: 10.1016/s0006-291x(86)80352-7. [DOI] [PubMed] [Google Scholar]
- Mencher J. R., Alford J. A. Purification and characterization of the lipase of Pseudomonas fragi. J Gen Microbiol. 1967 Sep;48(3):317–328. doi: 10.1099/00221287-48-3-317. [DOI] [PubMed] [Google Scholar]
- Moulin A., Fourneron J. D., Piéroni G., Verger R. Interface-mediated inactivation of pancreatic lipase by a water-reactive compound: 2-sulfobenzoic cyclic anhydride. Biochemistry. 1989 Jul 25;28(15):6340–6346. doi: 10.1021/bi00441a029. [DOI] [PubMed] [Google Scholar]
- Peled N., Krenz M. C. A new assay of microbial lipases with emulsified trioleoyl glycerol. Anal Biochem. 1981 Apr;112(2):219–222. doi: 10.1016/0003-2697(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Stuer W., Jaeger K. E., Winkler U. K. Purification of extracellular lipase from Pseudomonas aeruginosa. J Bacteriol. 1986 Dec;168(3):1070–1074. doi: 10.1128/jb.168.3.1070-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Oikawa T., Hirano K., Inukai T. Purification, crystallization and properties of triacylglycerol lipase from Pseudomonas fluorescens. Biochim Biophys Acta. 1977 Sep 28;488(3):353–358. doi: 10.1016/0005-2760(77)90194-1. [DOI] [PubMed] [Google Scholar]
- Verger R., De Haas G. H. Enzyme reactions in a membrane model. 1. A new technique to study enzyme reactions in monolayers. Chem Phys Lipids. 1973 Feb;10(2):127–136. doi: 10.1016/0009-3084(73)90009-1. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., D'Arcy A., Hunziker W. Structure of human pancreatic lipase. Nature. 1990 Feb 22;343(6260):771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- Winkler U. K., Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979 Jun;138(3):663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]