Abstract
Skn-1 is a maternally expressed transcription factor that specifies the fate of certain blastomeres early in the development of Caenorhabditis elegans. This transcription factor contains a basic region, but it binds to DNA as a monomer. Because other transcription factors containing basic regions bind as dimers, this finding implied that Skn represents a new DNA recognition motif. It has been proposed that the basic region helix of Skn is stabilized for binding by tertiary contacts to other parts of the protein. We have tested this proposal by carrying out circular dichroism (CD) and NMR experiments on the Skn domain and five truncated proteins. Our results have shown that the basic region of Skn is unstructured in solution and does not contact other parts of the protein; like other basic region peptides, it folds into a helix only upon binding specifically to DNA. However, there is a stably folded helical module in the Skn domain, and one of the helices in this module terminates immediately before the start of the basic region. This pre-organized helix contains a surface rich in basic amino acids, and we propose that this helix contacts the DNA distal to the basic region proper, providing an extra long helical recognition surface which helps to stabilize monomeric binding. Homology between the Skn domain and several basic-region leucine zipper (bZIP) domains raises the possibility that the affinity and perhaps the specificity of DNA binding by bZIP proteins can be modulated by incorporating a stably folded helical segment that contacts the DNA just below the basic region proper.
In 1992 a transcription factor that represents the first member of a new family of transcription factors was identified in Caenorhabditis elegans (1). This transcription factor, Skn-1, is a maternally expressed protein that acts at an early stage in the development of C. elegans embryos, specifying the formation of the pharynx and gut. The amino acid sequence of the binding domain revealed a basic region peptide at the C terminus and a 5-amino acid “homeodomain arm” at the N terminus, with a long, uncharacterized stretch of amino acids between these two elements. Basic region peptides are DNA recognition elements found in the basic-region leucine zipper (bZIP) and basic helix–loop–helix (bHLH) families of transcription factors. All known bZIP and bHLH proteins contain dimerization domains and bind to DNA as dimers (2, 3). Skn-1, however, did not appear to contain a dimerization domain. Furthermore, the consensus recognition site was identified as ATTGTCAT, which contains only a single basic region half site. Biochemical experiments have established that Skn-1 binds to DNA as a monomer (4).
It is worth noting that there have been numerous efforts to design monomeric proteins containing basic region recognition elements. Some of these attempts were made primarily to shed light on the factors involved in bZIP–DNA recognition (5, 6), but some have been made with a view toward learning how to utilize basic regions in the design of new DNA binders (7–9). Although several synthetic approaches have shown promise, attempts to design a monomeric basic region protein that can be expressed in cells have not been successful. Skn-1 is a natural blueprint for how to utilize basic region recognition elements in the design of monomeric DNA binding proteins (10).
The amino acid sequence of the DNA binding domain of Skn-1 yields clues about how this protein achieves monomeric binding. For example, the N-terminal “homeodomain arm” is identical to the N-terminal arm in the homeodomain protein, Antennapedia (4). Homeodomains consist of a stably folded three-helix domain, which binds in the major groove, and a flexible N-terminal arm, which binds in the minor groove (11). There is biochemical evidence that the N-terminal arm of the Skn domain likewise binds in the minor groove (4). Hence, although Skn presents only a single basic region to the DNA, contacts from the N-terminal arm help stabilize the complex.
Analysis of the amino acid sequence of the internal region did not yield useful information about its role in DNA binding. Blackwell et al. (4) suggested that this internal region is helical and forms tertiary contacts to the basic region, preorganizing it for binding and also helping to orient it in the major groove. According to this model, the Skn domain is essentially a helix–turn–helix protein which contains a stably folded recognition helix. Because all known basic region recognition helices are unfolded in solution and form stable helices only upon binding to DNA, the existence of a pre-organized basic region in Skn-1 would have interesting implications. Therefore, we decided to determine the solution conformation of the basic region in the Skn domain.
In addition, we thought that direct information about the structure of the internal region of the Skn domain might provide a basis for proposing a model for its role in DNA binding. Because the internal region of Skn bears significant homology to parts of several other transcription factors, including CNC, NF-E2, and NRF1, structural information on Skn might help in evaluating structure–function relationships in these proteins as well (4).
Biophysical studies on the Skn domain and five truncated proteins are reported. Based on information about the solution structures of both the basic region and the internal region, we have proposed a role for the internal region of the Skn domain in DNA binding. Although a high resolution structure of Skn bound to DNA will ultimately be required for a full understanding of the complex, the information presented here should help guide the design of new experiments to probe Skn binding. The results may have implications for how to influence affinity and specificity in bZIP proteins.
MATERIALS AND METHODS
Construction of Plasmids.
The gene encoding the 85-amino acid Skn domain flanked by appropriate restriction enzyme sites was synthesized from 15 overlapping oligonucleotides and cloned into M13mp18. The Cys-70–Ser-70 mutation was introduced using the Kunkel method (12). After sequence verification, the SknOH gene containing the serine mutation was subcloned into pET3a (Novagen). The gene encoding the N-terminal truncated protein was produced by PCR amplification of the M13mp18-SknOH plasmid using the universal M13 sequencing primer and primer N5, which was designed to introduce an ATG start codon immediately before the first arginine codon in the Skn gene. The 3′ terminal truncated genes were produced by PCR amplification of the original SknOH gene using primer N5 as well as a synthetic primer designed to introduce a stop codon prematurely in the gene sequence. The sequences of the primers are as follows: M13 primer, 5′-GTAAACGACGGCCAGT; N5, 5′-CCAGGGAATTCGGCATATGCGTAAACGTGGTCGTCAGTC; C80, 5′-GGGGGCGGATCCCTATTATTTGTCGTGACGGTCGGTA; C77, 5′-GGGGGCGGATCCCTATTAACGGTCGGTACGACGCTGACG; C74, 5′-GGGGGCGGATCCCTATTAACGACGCTGACGGGAGGT; C71, 5′-GGGGCGGATCCCTATTAACGGGAGGTACGAGCAGC; C64, 5′-GGGGGCGGATCCCTATTATTTGTTTTTACCACGACGACG.
After cloning into pET3a, the gene sequences were verified by Sanger dideoxy sequencing using [α-32P]ATP or by automated fluorescent DNA sequencing (SeqWright, LLC, Houston, TX).
Protein Purification.
The Skn genes were expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen). The cell lysate from a 500-ml culture in 10 mM Tris⋅HCl (pH 8.0) was centrifuged at 4°C for 20 min at 55,000 × g. The supernatant was passed over a 25 ml CM-Sephadex column equilibrated in 0.05 M phosphate, 0.05 M NaCl (pH 7.0). The protein was eluted with a sodium chloride step gradient. Fractions containing Skn were identified by monitoring the tyrosine absorbance at 274 nm. After concentration in a Centricon MWCO 3K filtration unit (Amicon), proteins were desalted on a PD-10 column (Pharmacia). All proteins were purified to >95% homogeneity (13). The compositions were verified by amino acid analysis (Princeton Synthesis Facility).
Protein concentrations were estimated from the tyrosine absorbance at 274 nm using a ɛmax of 2,800 M−1⋅cm−1 for N5-C85 and 1,400 M−1⋅cm−1 for the C-terminal truncated proteins. Solvent denaturation did not significantly alter the absorbance at 274 nm.
Electrophoretic Mobility Shift Assays (EMSAs).
An 18-base oligonucleotide (SKN18) containing the Skn binding site, 5′-GCTACATTGTCATCCCTC, and a control oligonucleotide (NS18) with the sequence 5′-GTACCACTGGCGGTGATC, were 5′-labeled with [γ-32P]ATP and annealed with a 5-fold excess of their complements. A typical binding mixture contained the following in a total volume of 10 μl: 80 mM Tris⋅HCl (pH 7.2), 1 mM EDTA, 10 mM KCl, 1 mM DTT, 0.2% Nonidet P-40, 10% glycerol, 2 mg/ml BSA, 1% bromophenol blue dye, 1000 cpm labeled DNA (>106 cpm/pm), and protein. Protein concentrations were varied 2-fold after the appropriate concentration ranges were identified in preliminary experiments. Binding mixtures were equilibrated for 1 h at 4°C before being loaded on a 10% TE gel (10 mM Tris/1 mM EDTA, pH 7.4), which was run at 100 V at 4°C for 3 h.
Circular Dichroism (CD) Studies.
CD experiments were performed on an Aviv Associates (Lakewood, NJ) model 62DS spectrometer using a 0.1-cm cuvette (Helma, Jamaica, NY). Samples contained 15 μM protein and/or DNA in 0.1 M NaCl and 0.05 M phosphate (pH 7.0).
Wavelength scans were recorded at 25°C with a step size of 0.5 nm and 1-sec averaging time. Fractional helicities were calculated using the method of Chen et al. (14). Thermal melts were performed with a 2-min equilibration and a 1-min averaging time for each temperature point. The temperature was changed in steps of 1°C/min. Melting temperatures were determined from the minimum of the first derivative of the molar ellipticity at 222 nm with respect to the reciprocal of the temperature (15).
NMR Studies.
NMR samples were prepared by dialyzing purified protein against NMR buffer (0.01 M sodium phosphate/0.25 M NaCl, pH 7.0) and then concentrating the dialysate to 1.0 mM. Double-quantum, total correlation spectroscopy, and nuclear Overhauser effect spectroscopy spectra were acquired on a 600 MHz Unity/INOVA (Varian, Palo Alto, CA) NMR spectrometer at 25°C and processed and analyzed using nmrpipe (16) and nmrview (17) softwares.
RESULTS AND DISCUSSION
Expression and Purification of the Skn Domain.
The gene encoding the 85-amino acid Skn domain was synthesized from 15 overlapping oligonucleotides using codons that are abundant in genes encoding highly expressed E. coli proteins. To facilitate spectroscopic studies, Cys-70 in the basic region was switched to a serine to make the addition of reducing agents such as DTT unnecessary. Although a homology alignment with known basic region peptides suggests that the cysteine side chain directly contacts the DNA, work on several bZIP proteins has shown that cysteine and serine are functionally interchangeable at this position (18, 19). The synthetic gene was cloned into pET3a, a T7 expression system (20), and the protein was expressed in BL21(DE3)pLysS cells. SDS/PAGE gel analysis of the cell lysate from an induced culture showed small amounts of a foreign protein with an apparent molecular mass of 12.0 kDa (21). The ability of the induced protein to bind to DNA was evaluated by running a gel shift assay on the crude cell lysates from both the induced and uninduced cultures. Only the induced culture was able to retard a radiolabeled duplex, SKN18, containing the Skn recognition site, confirming that the protein produced upon induction from the synthetic gene contained the expected DNA binding activity.
The yield of the SknOH protein was disappointing (≈1 mg/liter); however, protein expression in bacterial cells is sensitive to a wide variety of factors, including the amino acid sequence at both the N and the C termini (22, 23). Because the first four amino acids in the Skn domain are not part of the amino terminal arm, and there is no evidence that they play a functional role in binding, we re-engineered the SknOH gene to remove these amino acids. This N-terminal truncated protein N5-C85 (Fig. 1b), which contains an arginine immediatly after the initiating methionine, was expressed at much higher levels than SknOH itself. N5-C85 was purified by passage over CM-sephadex (Fig. 1c). A total of 15 mg/liter of protein purified to >95% homogeneity could be obtained after a single column.
Figure 1.
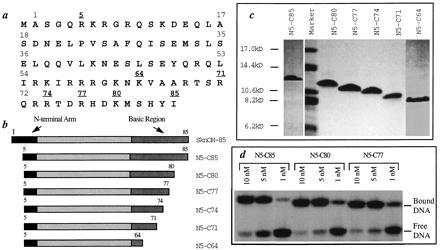
(a) The amino acid sequence of the SknOH protein. (b) Schematic of the six Skn proteins. (c) Coomassie blue-stained gel of the purified SknOH proteins. (d) Gel shift data for N5-C85, N5-C80, and N5-C77 bound to the SKN18 recognition duplex.
An EMSA under low salt conditions showed that purified N5-C85 has a Kd of ≈2.5 nM for the SKN18 duplex (Fig. 1d). The protein did not bind to the nonspecific control duplex, NS18, even at protein concentrations up to 12 μM (data not shown). Furthermore, a competition experiment in which unlabeled NS18 was added to the specific DNA complex showed that the specific complex remained almost fully bound in the presence of 10 μM NS18. Hence, the protein expressed in bacterial cells from the synthetic gene binds specifically to the Skn-1 recognition sequence with a dissociation constant comparable to that reported for the wild-type protein (4).
CD Studies on N5-C85.
Both homeodomain proteins and bZIP proteins utilize a recognition helix that makes specific contacts to the major groove upon binding to DNA. Homeodomain motifs are stably folded globular structures that are predominantly helical in solution (24); although the recognition helices can elongate, there are no major conformational transitions upon binding to DNA (25). In contrast, the basic region peptides in bZIP proteins undergo a coil-to-helix transition upon DNA binding (26–28). To shed more light on how the Skn domain compares to these other families of transcription factors, we carried out CD studies on N5-C85 in both the absence and presence of DNA.
The CD spectrum of N5-C85 in solution exhibits minima at 206 nm and 222 nm, indicating at least partial α-helicity (Fig. 2a). The α-helical content of the protein was estimated to be ≈30% based on the molar ellipticity at 222 nm (14). This estimate corresponds to ≈25 residues in a helical conformation. The calculated number of helical residues is only an approximation, but it serves as a reference for comparing other proteins (see below).
Figure 2.
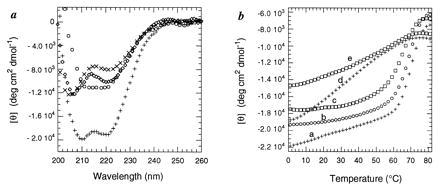
(a) CD curves of N5-C85 alone (×); difference CD curves (complex-free DNA) of N5-C85 with the Skn recognition duplex (+), and of N5-C85 with the nonspecific control duplex (⋄); ○, difference between the complex and the sum of the spectra of the free protein and the free SKN18 duplex. (b) Thermal denaturation curves of N5-C85 (curve a, +), N5-C77 (curve b, ○), and N5-C74 (curve c, □) in the presence of SKN18; N5-C85 in the presence of NS18 (curve d, +); and N5-C74 in the presence of NS18 (curve e, □). Although N5-C74 has a lower melting temperature and a slightly broader transition than the other proteins, a comparison of curves c and e confirms that it binds specifically to the SKN18 site.
Upon adding SKN18, the helical content of the protein increased significantly (Fig. 2a). The minimum at 222 nm decreased substantially, and the minimum at 206 nm shifted to 208 nm and also decreased. Although alterations in the DNA conformation upon protein binding can affect estimates of the number of induced helical residues (Table 1), which are based on curves in which the free DNA is subtracted, the magnitude of the change observed when N5-C85 binds provides unambiguous evidence for a substantial helical transition.
Table 1.
The fractional helicity fH (%) and the corresponding number of helical residues (n) of each Skn protein and the corresponding complexes
| Protein | Free protein
|
Complex
|
Tm, °C | ||
|---|---|---|---|---|---|
| fH, % | n | fH, % | n | ||
| N5-C85 | 30.0 | 25.0 | 60.0 | 49.0 | 70.9 |
| N5-C80 | 32.0 | 25.0 | 63.0 | 49.0 | 72.9 |
| N5-C77 | 31.0 | 23.0 | 56.0 | 43.0 | 70.9 |
| N5-C74 | 32.0 | 23.0 | 51.0 | 36.0 | 67.0 |
| N5-C71 | 37.0 | 25.0 | — | — | — |
| N5-C64 | 41.0 | 27.0 | — | — | — |
Three helical segments were assumed for the free proteins, and four were assumed for the bound proteins. Although changing the number of helices would change the calculated numbers slightly, conclusions drawn from the comparisons between the different proteins should be reliable as long as the proteins in question contain the same number of helical segments. The melting temperatures (Tm) of the specific complexes are shown in the last column (26).
The nonspecific DNA duplex did not have the same dramatic effect on the conformation of the Skn domain, although a moderate increase in helicity was observed (Fig. 2a). The increase in helical content in the protein in the presence of NS18 may reflect stabilization of a partially folded “nascent” basic region helix by nonspecific electrostatic interactions. Consistent with this hypothesis, polyanionic heparin fragments also induce a small increase in helicity in N5-C85. Thermal melts of the two different DNA complexes show that N5-C85 bound to SKN18 unfolds cooperatively (Tm ≈ 71°C) consistent with the melting of a single, specific complex, while N5-C85 bound to NS18 unfolds over a broad temperature range (Fig. 2b, curves a and d).
Synthesis and Evaluation of Truncated Proteins.
The basic regions of bZIP domains are known to fold on binding specifically to DNA (26–28). The sequence homology between the basic region of Skn and the basic regions of various bZIP domains suggests that the folding transition that occurs in N5-C85 in the presence of the specific DNA recognition site involves the C-terminal basic region. To test this hypothesis, and to simultaneously determine the minimum functional length of the Skn domain, we constructed a series of C-terminal truncation mutants and evaluated their behavior in the presence and absence of DNA. We reasoned that if the basic region in the free protein is not helical and does not interact with the rest of the protein, there should be no significant change in the estimated number of helical residues in the Skn domain as the C terminus is truncated. Moreover, if the basic region does, in fact, undergo a coil-to-helix transition upon binding to DNA, the number of induced helical residues might change as the protein becomes shorter.
Truncation mutants missing 5, 8, 11, 14, and 21 residues from the C terminus were engineered, expressed, and purified by passage over CM-sephadex (Fig. 1 b and c). Electrophoretic mobility shift analysis showed that the N5-C80 and N5-C77 proteins bind to the consensus recognition site with dissociation constants comparable to the full-length protein (Fig. 1d). Furthermore, neither protein binds to the nonspecific duplex even at 10 μM concentrations. Therefore, 8 amino acids can be removed from the C terminus of the protein without significantly altering the DNA binding behavior. In contrast, N5-C74 binds four times more weakly than the full-length protein (Kd ≈ 10 nM), and there is a reduction in the binding specificity. Whereas the full-length protein remains almost fully bound to the Skn site in the presence of 10 μM NS18, these same concentrations of NS18 displace N5-C74 from the Skn site. Nevertheless, N5-C74 still shows a distinct preference for SKN18 over NS18, suggesting that it contains most of the residues that determine specificity. N5-C71, which is 3 amino acids shorter than N5-C74, binds weakly (Kd ≈ 10−5 M) and nonspecifically to both DNA sequences. Because N5-C71 showed no specificity for DNA, we did not evaluate the DNA binding behavior of the shortest protein, N5-C64.
The structures of the five C-terminal truncated proteins were evaluated by CD spectroscopy using the same conditions as for N5-C85. The estimated numbers of helical residues in all five proteins were essentially identical to the number estimated for the full-length protein (Table 1).
Moreover, in the presence of SKN18, the three proteins that had been found to bind specifically by EMSA analysis (N5-C80, N5-C77, and N5-C74) showed large increases in helicity (Table 1) and cooperative melting transitions (Fig. 2b). Like the full-length protein, they did not melt cooperatively in the presence of NS18. A titration of N5-C77 with SKN18, showed that the induced helicity stops increasing once a 1:1 ratio of protein to DNA is achieved. This result is consistent with previous findings by Blackwell et al. (4) showing that the Skn domain binds to DNA as a monomer. N5-C71 did not increase in helicity in the presence of SKN18.
The results also show that number of helical residues that are induced upon DNA binding seems to correlate with the length of the C terminus. Although we have already noted that changes in the DNA conformation on binding can have large effects on the estimated number of induced helical residues, comparisons between the different complexes should be valid as long as the complexes bind to DNA in a similar mode. The EMSA studies indicate that N5-C80 and N5-C77 do bind in the same mode as the full-length protein.
We have drawn a number of conclusions from the studies described above. (i) The minimum length of the Skn domain for DNA binding is 73 amino acids, and the last eight residues of the C terminus do not play a significant role in DNA binding. (ii) Skn contains a helical domain that involves about one-third of the protein. (iii) The basic region of Skn does not interact with this helical domain because removing up to 21 residues has virtually no effect on the estimated number of helical residues. (iv) The Skn domain undergoes a major helical structural transition upon binding to DNA. (v) Because the number of helical residues induced upon binding to DNA appears to correlate with the length of the C terminus, it is likely that the observed transition involves folding of the basic region into a helix. This conclusion is supported by analogy to the basic regions found in other transcription factors, which undergo a coil-to-helix transition upon binding to DNA.
NMR Studies.
CD provides information about the average number of helical residues in a protein, but does not provide detailed information about structure. We have undertaken NMR studies on some of the proteins described above to obtain more detailed information on the Skn domain. NMR results pertaining to the present discussion are presented below.
One-dimensional proton NMR spectra of the amide regions of N5-C77 and N5-C64 at 25°C are shown in Fig. 3a. N5-C77 binds specifically to the Skn site and contains a fully functional basic region; N5-C64 lacks almost the entire basic region. The NMR spectra for both proteins show sharp lines for most of the resonances, with good dispersion, and only a single set of peaks. The amide resonances downfield of ≈8.5 ppm provide graphic evidence that both N5-C77 and N5-C64 contain a stably folded domain. The downfield shifted amide signals are virtually identical for N5-C77 and N5-C64 even though the latter is 13 residues shorter. Hence, removing the basic region does not alter the structure of the folded domain. The NMR data confirm the CD results that the basic region does not interact with the rest of the protein.
Figure 3.
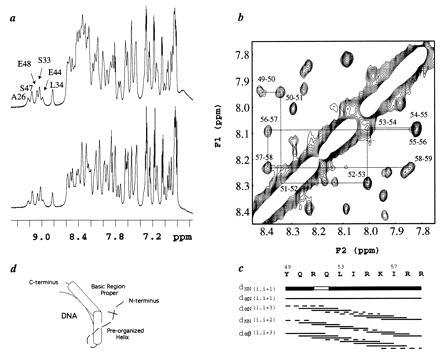
(a) 1H NMR spectra of the amide regions of N5-C77 (Upper) and N5-C64 (Lower). (b) A portion of the nuclear Overhauser effect spectroscopy (NOESY) spectrum for N5-C77 showing the dNN connectivities for residues 49–59. (c) Summary of sequential and medium range connectivities for residues 49–59. The gap in the dNN (i, i+1) connectivity and the dashed lines in the medium-range connectivities also indicate overlap. (d) A schematic showing the bipartite recognition helix of Skn in DNA binding. The tertiary structure of the folded domain has not been sufficiently defined to include in the model.
Sequential assignments have been made for residues L16–R59. In addition, several of the resonances in the basic region of N5-C77 have been assigned, including N63, V65, A66, A67, T69, S70, and T75. These basic region residues have 1H chemical shifts very close to the random coil values, supporting the CD studies which suggested that the basic region is unstructured in solution. In contrast, the internal region of the Skn domain is largely helical. Three stably folded helical segments have been identified between residues 27–59 on the basis of strong sequential amide-amide NOEs, the presence of αN(i, i+3) and αβ(i, i+3) nuclear Overhauser effects (NOEs), and in some cases, the presence of αN(i, i+4) NOEs. The most striking finding is that one of the helices spans residues 49–59, thus extending right up to the start of the basic region (Fig. 3 b and c). Structure prediction algorithms did not identify this region as helical (4).
Skn Uses a Bipartite Recognition Helix in DNA Binding.
The big question regarding the binding of Skn to DNA is how it is able to bind to DNA as a monomer. Previous results had implicated the basic region and the N-terminal arm in binding; however, the role of the internal region was unclear. It was suggested that the internal region promotes monomeric binding by stabilizing the basic region via tertiary contacts. Our results show that this is not the case: the basic region is unstructured in solution and does not interact with the rest of the protein. Skn does, however, contain a stably folded helical domain located between the C-terminal basic region and the N-terminal arm. If this domain does not interact directly with the basic region, what is its role?
One possibility is that the folded domain may function primarily as a scaffold which orients the N-terminal arm and the C-terminal basic region for binding, but does not itself contact the DNA. The importance of having a scaffold that properly orients DNA binding elements is clear both from first principles (29) and from studies on other transcription factors (8).
It is also possible, however, that the internal region of Skn plays a dual role, helping to orient the C-terminal basic region and the N-terminal arm, and also contributing directly to binding by establishing contacts to the DNA. To understand how Skn achieves monomeric binding, it is critical to distinguish between these different models for the role of the internal region. We believe that the structural information reported above provides compelling support for the second model. Our arguments are presented below.
Crystal structures on the bZIP domains of GCN4 and Fos-Jun bound to DNA are available (30–32). The boundaries of the basic regions in these proteins can be defined by the residues which contact the DNA. If we assume that the basic region of Skn binds to DNA in the same way as the basic regions of these other proteins, then the basic region proper of Skn extends from about R60 to R77. Several lines of evidence support the assumption that the Skn basic region binds like these other basic regions. (i) The Skn basic region behaves like these other basic regions in that it undergoes a coil-to-helix transition upon DNA binding. (ii) The Skn basic region contains the same recognition amino acids and has the same half-site specificity as these other basic regions. (iii) Studies carried out in our laboratory show that mutations in several residues predicted to contact the DNA based on homology decrease the affinity and specificity of DNA binding (data not shown).
Our NMR studies have identified a stably folded helix (Fig. 3 b and c) that, in solution, terminates at approximately R60, which is at the start of the unstructured basic region proper. This pre-organized helix contains a surface that is rich in basic amino acids and a surface that is rich in hydrophobic amino acids. Upon binding to DNA, folding of the unstructured basic region would produce a continuous helix consisting of the basic region proper and the “pre-organized” helical segment preceding it. If the basic region proper were docked into DNA with the recognition amino acids making the appropriate contacts, the basic surface of the pre-organized helical segment would be oriented toward the DNA and could interact with the DNA. Monomeric binding may, therefore, be stabilized not only by contacts from the N-terminal arm and the basic region proper to the DNA, but also by contacts from the pre-organized helix which immediately adjoins the basic region helix (Fig. 3d).
The proposal that the Skn domain binds to DNA using a bipartite recognition helix that is a composite of a basic region helix and a pre-organized helix is consistent with ethylation interference data showing that Skn makes contacts to the DNA backbone several residues beyond the TCAT basic region half site (4). It is also consistent with evidence suggesting that the binding site preferences of Skn extend beyond the basic region half site (4). However, the model requires that there be a kink or a hinge between the two helical segments. Otherwise, the composite recognition helix would diverge from the DNA below the basic region proper. It is notable that there is a glycine near the beginning of the Skn basic region. This glycine is conserved in a number of basic region transcription factors which contain significant homology to the internal region of Skn (4, 33). Glycine is an amino acid that confers flexibility and does not favor propagation of a helix. This glycine may play a role in permitting the extra-long recognition helix to bend sufficiently to maintain contact with the DNA.
Implications for Other Transcription Factors.
The striking homology between the internal region of the Skn domain and regions of several other transcription factors which contain basic regions, including CNC, NF-E2, and NRF1, raises the possibility that these proteins also contain a stably folded helix which adjoins the basic region and interacts with DNA (4, 33). Many of these transcription factors belong to the bZIP family. It has been noted by many that a large number of bZIP proteins contain the same recognition amino acids and have similar half-site specificity, raising questions about how diversity can be achieved in DNA binding among proteins that contain very similar basic regions (34, 35). Mechanisms for achieving diversity in binding have focused on how heterodimerization and amino acids at the fork region of the leucine zipper influence binding specificity (34, 35). Our work on the Skn domain raises the possibility that including a pre-organized helical region adjacent to the basic region may be another way of modulating the affinity and/or specificity of basic region-containing proteins. In fact, studies on NF-E2, which contains a basic region homologous to Fos-Jun, have shown that it recognizes an extended binding site compared with Fos-Jun (33). The results presented above should help guide future biochemical investigations of these unusual bZIP proteins as well as on Skn-1.
ABBREVIATIONS
- bZIP
basic-region leucine zipper
- CD
circular dichroism
- EMSA
electrophoretic mobility shift assay
References
- 1.Bowerman B, Eaton B A, Priess J R. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 2.Hope I A, Struhl K. EBMO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park C, Campbell J L, Goddard W A I. J Am Chem Soc. 1996;118:4235–4239. [Google Scholar]
- 4.Blackwell T K, Bowerman B, Priess J R, Weintraub H. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- 5.O’Neil K, Hoess R H, DeGrado W F. Science. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 6.Talanian R V, McKnight C J, Kim P S. Science. 1990;249:769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 7.Park C, Campbell J L, Goddard W A I. J Am Chem Soc. 1995;117:6287–6291. [Google Scholar]
- 8.Cuenoud B, Schepartz A. Proc Natl Acad Sci USA. 1993;90:1154–1159. doi: 10.1073/pnas.90.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno M, Sawada M, Makino K, Morri T. J Am Chem Soc. 1994;116:11137–11138. [Google Scholar]
- 10.Deng T, Karin M. Proc Natl Acad Sci USA. 1992;89:8572–8576. doi: 10.1073/pnas.89.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ades S E, Sauer R T. Biochemistry. 1995;34:14601–14608. doi: 10.1021/bi00044a040. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y-H, Yang J T, Chau K H. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 15.Cantor C R, Schimmel P R. Biophysical Chemistry. Vol. 3. San Francisco: Freeman; 1980. pp. 1130–1133. [Google Scholar]
- 16.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 17.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 18.Abate C, Patel L, Rauscher F J, III, Curran T. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 19.Sera T, Schultz P G. Proc Natl Acad Sci USA. 1996;93:2920–2925. doi: 10.1073/pnas.93.7.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studier F W, Moffat B A. J Mol Biol. 1986;189:113–117. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Abate C, Luk D, Gentz R, Rauscher F, III, Curran T. Proc Natl Acad Sci USA. 1990;87:1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirel P-H, Schmitter J-M, Dessen P, Fayat G, Blanquet S. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 24.Shang Z, Isaac V E, Li H, Patel L, Catron K M, Curran T, Montelione G, Abate C. Proc Natl Acad Sci USA. 1994;91:8373–8377. doi: 10.1073/pnas.91.18.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsao D H H, Gruschus J M, Wang L-H, Nirenberg M, Ferretti J A. Biochemistry. 1994;33:15053–15060. doi: 10.1021/bi00254a014. [DOI] [PubMed] [Google Scholar]
- 26.Weiss M A, Ellenberger T, Wobbe C R, Lee J P, Harrison S C, Struhl K. Nature (London) 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 27.Patel L, Abate C, Curran T. Nature (London) 1990;347:572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- 28.O’Neil K T, Shuman J D, Ampe C, DeGrado W F. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- 29.Jencks W P. Proc Natl Acad Sci USA. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller W, Konig P, Richmond T J. J Mol Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 31.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 32.Glover J N M, Harrison S C. Nature (London) 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 33.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Tzamarias D, Ellenberger T, Harrison S. Proc Natl Acad Sci USA. 1993;90:4513–4517. doi: 10.1073/pnas.90.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinson C R, Hai T, Boyd S M. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]


