Abstract
Bloom’s syndrome (BS) is an autosomal recessive disorder characterized by a strong cancer predisposition. The defining feature of BS is extreme genome instability. The gene mutated in Bloom’s syndrome, BLM, encodes a DNA helicase (BLM) of the RecQ family. BLM plays a role in homologous recombination; however, its exact function remains controversial. Mutations in the BLM cause hyperrecombination between sister chromatids and homologous chromosomes, indicating an anti-recombination role. Conversely, other data show that BLM is required for recombination. It was previously shown that in vitro BLM helicase promotes disruption of recombination intermediates, regression of stalled replication forks, and dissolution of double Holliday junctions. Here, we demonstrate two novel activities of BLM: disruption of the Rad51-ssDNA (single-stranded DNA) filament, an active species that promotes homologous recombination, and stimulation of DNA repair synthesis. Using in vitro reconstitution reactions, we analyzed how different biochemical activities of BLM contribute to its functions in homologous recombination.
[Keywords: Bloom’s syndrome, BLM helicase, homologous recombination, Rad51, RecQ, Rad54]
In humans, mutations in the BLM gene cause Bloom’s syndrome (BS), an autosomal recessive disorder whose clinical manifestations include proportional dwarfism, immunodeficiency, male infertility, and others (German 1993). A specific feature of BS is a greatly elevated incidence of cancer with exceptionally early onset. BS individuals are predisposed to develop most types of cancer, an unusual feature among cancer predisposition disorders. The hallmark of BS is a high degree of genome instability (Bachrati and Hickson 2003; Hickson 2003). Specifically, BS shows a greatly increased frequency of reciprocal sister chromatid exchanges (SCE) and exchanges between homologous chromosomes (German 1993). These data indicate that BLM suppresses homologous recombination (HR), the process responsible for chromosomal exchanges (Sonoda et al. 1999; Gonzalez-Barrera et al. 2003). However, recent genetic data revealed a more complex relationship between BLM and HR: BLM may suppress some recombination events and promote others (Adams et al. 2003).
In eukaryotes, HR has several crucial functions, including segregation of homologous chromosomes, propagation of genetic diversity, maintenance of telomeres, and repair of double-stranded DNA breaks (DSB) (Hoeijmakers 2001; West 2003; Krogh and Symington 2004; Whitby 2005; Agarwal et al. 2006; Sung and Klein 2006; Helleday et al. 2007). HR is performed by an assembly of specialized proteins, in which Rad51 plays a central role (Bianco et al. 1998). Rad51 forms a nucleoprotein filament on single-stranded DNA (ssDNA), which is generated by specialized exonucleases at the sites of DNA breaks. The filament possesses unique activities; it performs the search for homologous double-stranded DNA (dsDNA) sequences and promotes subsequent DNA strand exchange between ssDNA and homologous dsDNA sequences (Sung et al. 2003). DNA strand exchange results in formation of joint molecules (D-loops), in which the invading ssDNA serves as a primer and the homologous dsDNA as a template for DNA polymerase during DNA repair synthesis. The joint molecules continue down one of the two pathways (Allers and Lichten 2001; Hunter and Kleckner 2001). They either dissociate, leading to rejoining of the broken chromosome through synthesis-dependent strand annealing (SDSA) (Allers and Lichten 2001), or proceed through the capture of the second processed DNA end to produce Holliday junctions (Schwacha and Kleckner 1995; Cromie et al. 2006), which are later resolved by structure-specific endonucleases to produce crossover and noncrossover recombinants via the DSB repair (DSBR) mechanism (Pâques and Haber 1999). Whereas crossing over is essential for proper chromosome disjunction during meiosis, it may be detrimental during mitotic recombination due to loss of heterozygosity (LOH) and genome rearrangements. Consequently, mitotic recombination proceeds mainly through the SDSA mechanism producing noncrossover recombinants.
The product of the BLM gene is BLM helicase, a member of the highly conserved RecQ family, which is responsible for genome maintenance in all organisms from bacteria to humans (Wu and Hickson 2006). Consistent with its role in HR, BLM physically interacts with HR proteins Rad51 and Rad51D, as well as with several other proteins involved in DNA repair and DNA-damage signaling such as Mus81, MLH1, RPA, and ATM (Beamish et al. 2002; Sharma et al. 2006). BLM can also specifically recognize Holliday junctions and promote their branch migration in an ATPase-dependent manner (Karow et al. 2000). In vitro, BLM was shown to unwind D-loops (van Brabant et al. 2000; Bachrati et al. 2006), catalyze regression of model replication forks (Ralf et al. 2006), and resolve double Holliday junctions (Wu and Hickson 2003) by forming a complex with topoisomerase IIIα (Topo IIIα) and BLAP75 (Raynard et al. 2006; Wu et al. 2006).
Here we identified two novel biochemical activities of BLM. We found that it can disrupt the Rad51-ssDNA filament by dislodging human Rad51 (hRad51) protein from ssDNA in an ATPase-dependent manner, the activity consistent with suppression of HR at an early stage. We also demonstrated that BLM can stimulate DNA repair synthesis and thereby possibly promote HR at the late stages. Using in vitro reconstitution of HR reactions we analyzed how the biochemical activities of BLM can either promote or inhibit the SDSA mechanism of HR at different stages.
Results
BLM inhibits DNA strand exchange activity of Rad51
BLM helicase shares similarity in its biochemical properties with the yeast Srs2 helicase, which is known to dissociate the Rad51 filament (Krejci et al. 2003; Veaute et al. 2003). In addition, overexpression of Sgs1, the sole yeast RecQ ortholog, can suppress recombination defects of srs2Δ, indicating at least partial overlap in their functions (Mankouri et al. 2002; Ira et al. 2003). We hypothesized that BLM can dissociate the filament formed by hRad51 on ssDNA, in the same manner as Srs2. Here we tested this hypothesis.
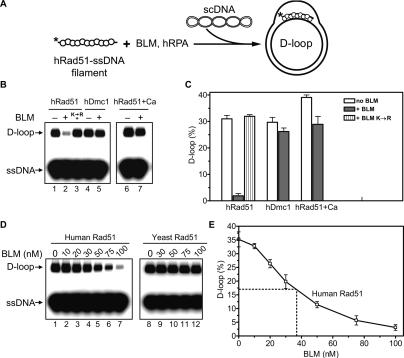
Since filament disruption would result in inhibition of DNA strand exchange, we first used the D-loop assay to test the effect of BLM helicase on DNA strand exchange promoted by hRad51. In this assay, hRad51 forms a nucleoprotein filament on ssDNA, which then catalyzes formation of joint molecules (D-loops) with pUC19 supercoiled DNA (scDNA) (Fig. 1A). The reaction also contained human RPA (hRPA), a ubiquitous eukaryotic ssDNA-binding protein involved in DNA repair, replication, and HR, which was added after hRad51-ssDNA filament formation. The presence of hRPA was expected to facilitate detection of hRad51 displacement from ssDNA by BLM: Previously, it was shown that yeast RPA by efficient binding to ssDNA prevents rapid reassembly of the yeast Rad51-ssDNA filament after its disruption by Srs2 (Krejci et al. 2003). As we showed previously, formation of an active hRad51-ssDNA filament requires Ca2+ (Bugreev and Mazin 2004). Therefore, to activate the hRad51-ssDNA filament, which was initially assembled in the presence of Mg2+ alone, calcium (2 mM) was added to the reaction mixture prior to addition of pUC19 scDNA. As expected, hRad51 efficiently promoted formation of D-loops (Fig. 1B, lane 1). However, addition of BLM to the hRad51-ssDNA filament prior to Ca2+ resulted in a strong inhibition of D-loop formation (Fig. 1B, lane 2). In contrast, we did not observe a significant inhibition when BLM was added to the filament after addition of Ca2+ (Fig. 1B, lane 7). The effect of the order of addition of BLM relative to Ca2+ indicated that the inhibition was mediated by BLM interaction with an inactive hRad51-ssDNA filament, presumably causing its disruption, not by melting of the D-loops. Ca2+ might prevent dislodging of hRad51 from ssDNA by BLM because it stabilizes the filament (Bugreev and Mazin 2004). The inhibition of D-loop formation showed dependence on the BLM concentration, with a half-inhibition (IC50) of ∼40 nM BLM (Fig. 1D,E). This BLM concentration was ∼75-fold lower than that of hRad51, indicating that BLM acts catalytically; e.g., by translocating along ssDNA. Consistent with this interpretation, the ATPase-deficient BLM mutant, K695R, which lacks DNA translocation and helicase activities, did not inhibit D-loop formation (Fig. 1B, lane 3). We found that inhibition of D-loop formation was specific for hRad51; BLM did not significantly inhibit DNA strand exchange activity of hDmc1, a meiotic hRad51 homolog (Fig. 1B [lanes 4,5], C), or yeast Rad51 protein (Fig. 1D, lanes 8–12). Conversely, inhibition of DNA pairing activity of hRad51 appeared to be specific for BLM, as RecQ1, another member of the human RecQ helicase family, in concentrations from 25 to 200 nM did not significantly inhibit Rad51-mediated D-loop formation under the tested conditions (data not shown). Overall, the data presented here suggest that the observed inhibition of D-loop formation is caused by disruption of the hRad51-ssDNA filament by BLM helicase.
Figure 1.
BLM inhibits DNA strand exchange activity of hRad51 protein by interaction with the hRad51-ssDNA filament. (A) The experimental scheme. The asterisk indicates the 32P label. (B) BLM (100 nM) was added to the hRad51 (lanes 1,2) or hDmc1 (lanes 4,5) nucleoprotein filaments assembled on ssDNA in the presence of 1 mM Mg2+. (Lane 3) In the control, BLM K695R (100 nM) was added to the hRad51-ssDNA nucleoprotein filament. D-loop formation was initiated by addition of 2 mM CaCl2 and pUC19 scDNA. (Lane 2) BLM inhibits D-loop formation, if added to the hRad51-ssDNA filament before Ca2+. In lanes 6 and 7, BLM (100 nM) was added to the hRad51-ssDNA filament formed in the presence of both 1 mM Mg2+ and 2 mM Ca2+. D-loop formation was initiated by addition of pUC19 scDNA. D-loops were analyzed by electrophoresis in a 1% agarose gel. (C) The results from B represented as a graph. (D) BLM in indicated concentrations was added to the nucleoprotein filaments formed by either hRad51 (lanes 1–7) or yeast Rad51 (lanes 8–12) proteins in the presence of Mg2+. D-loop formation was initiated by addition of either 2 mM CaCl2 (for hRad51) or 100 nM yeast Rad54 (for yeast Rad51) and pUC19 scDNA. (E) The results from D demonstrating inhibition of hRad51 by BLM are presented as a graph. Error bars in C and D indicate SEM.
BLM dismantles the hRad51-ssDNA filament
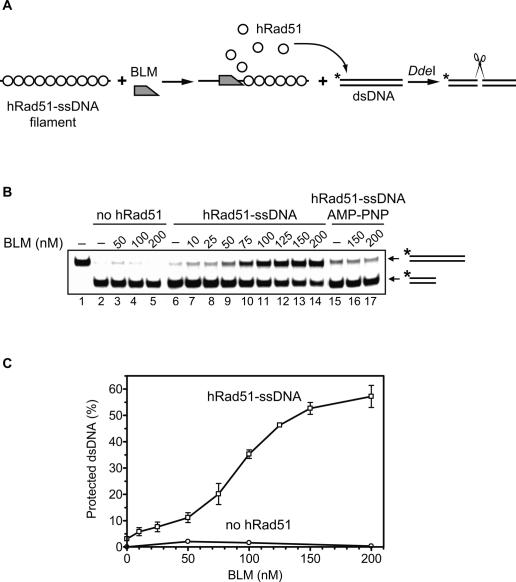
To demonstrate that BLM indeed displaces hRad51 from ssDNA, we developed a nuclease protection assay (Fig. 2A). Because hRad51 can bind both ssDNA and dsDNA, 32P-labeled dsDNA fragment was used as a trap for hRad51 that would be displaced from the filament. The dsDNA fragment contained a unique DdeI site, such that binding of hRad51 would protect the dsDNA from cleavage by DdeI. We demonstrated first that BLM or hRad51-ssDNA filament separately could not protect dsDNA against the cleavage (Fig. 2B [lanes 2–5,6], C). However, addition of BLM to the hRad51-ssDNA filament caused an inhibition of the DdeI cleavage in a BLM concentration-dependent manner, demonstrating the displacement of hRad51 from ssDNA (Fig. 2B [lanes 7–14], C). Cleavage protection was not observed in the presence of AMP-PNP, a nonhydrolysable ATP analog (Fig. 2B, lanes 15–17). This result argues against a hypothetical mechanism in which dsDNA would be protected from the cleavage by forming a nonspecific ternary complex with BLM and the hRad51-ssDNA filament.
Figure 2.
BLM promotes dissociation of the hRad51-ssDNA filament. (A) The experimental scheme of the restriction endonuclease protection assay. The asterisk indicates the 32P label. (B) The products of DNA cleavage were analyzed in a 10% polyacrylamide gel. BLM was added to the preformed hRad51-ssDNA filament, followed by addition of 32P-labeled dsDNA containing DdeI cleavage site. (Lanes 6–14) Protection against DdeI cleavage indicates the transfer of hRad51 from the nucleoprotein filament to the dsDNA probe (“hRad51-ssDNA”). In control, hRad51 was replaced by storage buffer (lanes 2–5, “no hRad51”), or reactions were carried out in the presence of AMP-PNP, a nonhydrolyzable ATP analog (lanes 15–17, “hRad51-ssDNA AMP-PNP”). Lane 1 shows migration of original dsDNA fragment noncleaved by DdeI. (C) The results from B presented as a graph. Error bars indicate SEM.
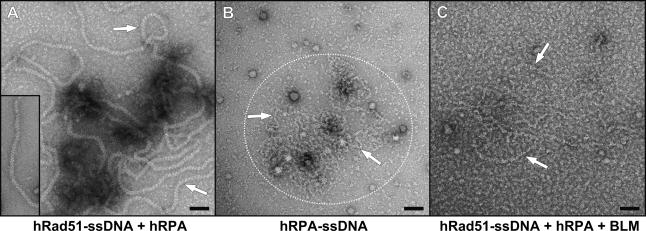
We also used electron microscopy to directly visualize the effect of BLM on the hRad51-ssDNA filament. After incubation of hRad51 with ssDNA, formation of the hRad51-ssDNA nucleoprotein filaments was detected (Fig. 3A). The filament was stable in the presence of hRPA (Fig. 3A), which formed distinct complexes with ssDNA in the absence of hRad51 (Fig. 3B). When BLM helicase was then added to the assembled hRad51-ssDNA complexes, hRad51 filaments could not be detected. Instead we observed the appearance of hRPA–ssDNA complexes (Fig. 3C). Thus, both the results of biochemical assays and electron microscopy demonstrated that BLM displaces hRad51 from ssDNA.
Figure 3.
Analysis of hRad51-ssDNA filament disruption by BLM using electron microscopy. (A) The hRad51-ssDNA filament (indicated by arrows) was formed and then mixed with hRPA. hRad51-ssDNA filament formed in the absence of hRPA is shown in the bottom left corner. (B) hRPA–ssDNA complexes (in the encircled area) are indicated by arrows. (C) The hRad51-ssDNA filament was formed and then mixed with BLM and hRPA. The reaction resulted in disruption of the filament and formation of hRPA–ssDNA complexes (indicated by arrows). Bar, 50 nm.
ATPase activity of BLM is strongly stimulated by ssDNA
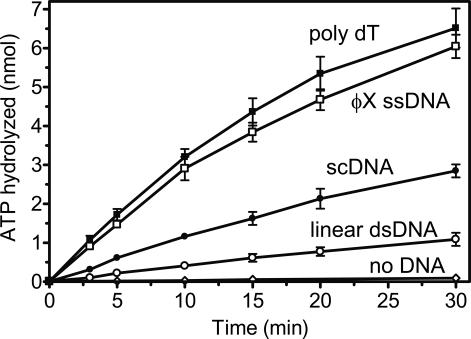
It is known that yeast Srs2, which displaces yeast Rad51 from ssDNA, has an ATPase activity that is strongly stimulated by ssDNA (Van Komen et al. 2003). Here we asked whether the ATPase activity of BLM shows similar specificity. Indeed, we found a strong ssDNA-dependent ATPase activity of BLM, which was approximately sixfold greater than its dsDNA-dependent ATPase (Fig. 4). In the previous report by Karow et al. (1997), preference of the BLM ATPase for ssDNA could have been missed because of the use of sheared salmon-sperm DNA substrates, which may contain some irregular structures like tailed or branched DNA that strongly induce the ATPase of BLM. Thus, the substrate specificity of BLM resembles that of yeast Srs2.
Figure 4.
ATPase activity of BLM is stimulated by ssDNA stronger than by linear or supercoiled (sc) dsDNA. ATP hydrolysis was carried out in the presence of poly dT (black squares), ϕX174 ssDNA (open squares), pUC19 scDNA (closed circles), or pUC19 linear dsDNA (open circles), or without DNA (diamonds). Error bars indicate SEM.
D-loop disruption by BLM
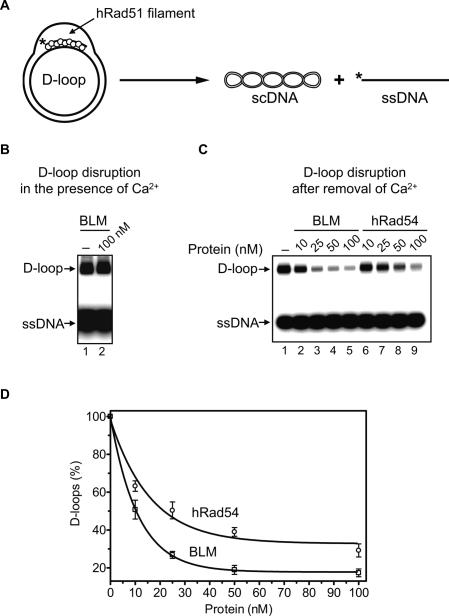
BLM can disrupt D-loops, key intermediates of HR (van Brabant et al. 2000; Bachrati et al. 2006). It was suggested that this activity of BLM may contribute to its anti-recombinase function. We recently found that hRad54 protein, owing to its ATP-dependent branch migration activity (Bugreev et al. 2006b; Mazina et al. 2007), can also cause dissociation of D-loops (Bugreev and Mazin 2007; Bugreev et al. 2007). However in contrast to BLM, hRad54 has no known anti-recombination function in vivo. We also found that the D-loop dissociation activity of hRad54 depends on the conformation of the hRad51 filament that remains associated with D-loops after their formation; i.e., the filament in an active ATP-bound form inhibits D-loop dissociation. Here we wanted to examine whether BLM can dissociate native (nondeproteinized) D-loops containing hRad51, which better mimic in vivo recombination intermediates than previously studied naked D-loops. We also wanted to determine whether the BLM ability to dissociate native D-loops is greater than that of hRad54, which would be consistent with its anti-recombination function.
We found that BLM efficiently dissociated D-loops after the hRad51 filament inactivation by removal of Ca2+ (Fig. 5C, lanes 2–5). In contrast, BLM was unable to dissociate native D-loops containing hRad51 when hRad51 was maintained in an active ATP-bound form in the presence of Ca2+ (Fig. 5B, lane 2). Since Ca2+ does not significantly inhibit dissociation of deproteinized D-loops by BLM, we attributed this inhibition to the effect of Ca2+ on the conformation of hRad51 filament (Bugreev and Mazin 2004). We then compared activities of BLM and hRad54 in dissociation of native D-loops after Ca2+ removal. Under the tested conditions, BLM showed only a slightly higher efficiency of D-loop dissociation than hRad54 (Fig. 5D).
Figure 5.
BLM promotes disruption of “native” D-loops containing hRad51. (A) The experimental scheme. The asterisk indicates the 32P label. (B) BLM cannot disrupt “native” D-loops in the presence of Ca2+. “Native” D-loops were formed by hRad51, and the reaction was initiated by addition of BLM (lane 2) or BLM storage buffer as a control (lane 1). The DNA products of D-loop disruption were analyzed by electrophoresis in a 1% agarose gel. (C) BLM (lanes 2–5) and hRad54 (lanes 6–9) can disrupt D-loops after Ca2+ removal. The protein concentrations are indicated above the gel. In lane 1, proteins were replaced by BLM storage buffer. (D) The data from C presented as a graph. Error bars indicate SEM.
Thus, the activity of BLM in D-loop dissociation closely parallels that of hRad54. We previously suggested that D-loop dissociation by hRad54 may represent a step leading to rejoining of the broken chromosome through the SDSA mechanism (Bugreev et al. 2007). Similarly, D-loop dissociation by BLM could also promote HR via the SDSA mechanism, rather than disrupt it. Genetic data also support the role of the Drosophila BLM ortholog (DmBLM) in the SDSA mechanism (Adams et al. 2003).
Stimulation of D-loop extension by BLM
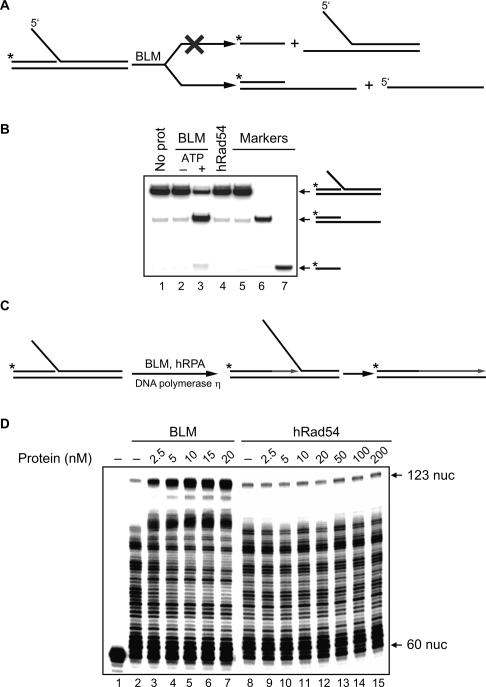
Sekelsky and coworkers (Adams et al. 2003) reported that BLM mutants in Drosophila both reduce the frequency of the SDSA-mediated HR and cause a decrease in the length of DNA synthesis tracts during the SDSA repair synthesis. They suggested that BLM may stimulate DNA repair synthesis by unwinding the DNA template ahead of the replication fork. Here, we wished to test the validity of this hypothesis for human proteins: BLM and DNA polymerase η. DNA polymerase η was chosen because of its reported implication in DSBR (Kawamoto et al. 2005; McIlwraith et al. 2005).
First, we tested the specificity of BLM in unwinding a model replication fork that mimicked the DNA joint molecule generated by ssDNA invasion into the homologous duplex DNA (Fig. 6A). The substrate was produced by annealing of synthetic oligonucleotides. Potentially, in this substrate, BLM helicase could unwind the displaced strand, the primer, or both. Using a 32P- labeled substrate, we found that, under the tested conditions, BLM was significantly more efficient in unwinding the displaced DNA strand (Fig. 6B, lane 3). Thus, the specificity of BLM helicase appeared consistent with its proposed role in unwinding the DNA template in front of the fork. In contrast, as expected, hRad54, which lacks DNA helicase activity, did not unwind this model replication fork (Fig. 6B, lane 4).
Figure 6.
BLM promotes unwinding of a model replication fork and stimulates primer extension by DNA polymerase η. (A) The experimental scheme. The asterisk indicates the 32P label. (B) BLM shows a preference in unwinding of the displaced DNA strand versus the primer. The DNA products were analyzed by electrophoresis in a 10% PAGE. The reactions were initiated by addition of BLM (10 nM) in the absence (lane 2) or presence (lane 3) of ATP. In controls, BLM was replaced by hRad54 (50 nM) (lane 4) or BLM storage buffer (lane 1). Migration of DNA markers is shown in lanes 5–7. (C) Schematic representation of replication fork extention by DNA polymerase η. The asterisk indicates the 32P label. (D) Effect of BLM (lanes 2–7) and hRad54 (lanes 8–15) in indicated concentrations on replication fork extension by DNA polymerase η. Lane 1 shows migration of a nonextended primer. The products of DNA synthesis were analyzed by denaturing PAGE.
We next examined the effect of BLM on DNA synthesis that was carried out by DNA polymerase η on the model replication fork (Fig. 6C). The results show that BLM stimulated DNA synthesis by DNA polymerase η (Fig. 6D, lanes 2–7). In contrast, hRad54 did not show the stimulatory effect on the DNA synthesis (Fig. 6D, lanes 8–15).
Discussion
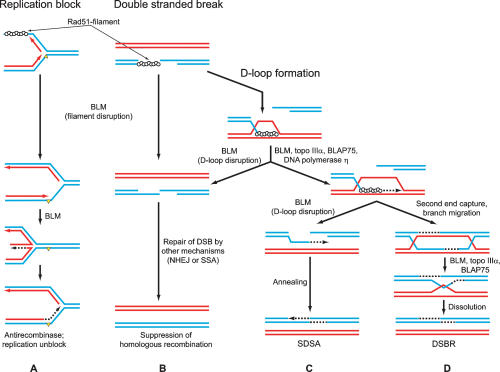
Although involvement of BLM in HR is well established, its precise function remains unknown. In this study, we discovered that BLM helicase can efficiently displace hRad51 protein from ssDNA, thereby disrupting the filament that catalyzes the essential initial steps of HR. We also found that BLM can stimulate DNA repair synthesis in vitro. These and previous data indicate that BLM may both suppress and stimulate the process of HR at different stages (Fig. 7).
Figure 7.
The role of BLM helicase in HR and in stalled replication fork repair. (A) By disruption of the Rad51-filament (top) BLM suppresses HR and promotes alternative repair mechanisms such as “template switching,” which involves replication fork regression (formation of the “chicken foot” structures) (bottom). (B) BLM disrupts Rad51 presynaptic filament, or D-loops, when HR cannot proceed further, preventing formation of potentially toxic intermediates and channeling DSBR into other pathways. (C) In D-loops, BLM stimulates primer extension by DNA polymerase η by unwinding DNA template ahead of the DNA polymerase. (Top) BLM-associated proteins, Topo IIIα and BLAP75, may facilitate this process by removing topological constraints that arise during BLM unwinding. (Bottom) BLM, by promoting D-loop disruption, channels DNA repair process into the SDSA pathway. (D) BLM–Topo IIIα–BLAP75 complex promotes resolution of double Holliday junctions, preventing mitotic crossing over.
Even though HR is indispensable for repair of damaged DNA and maintenance of genome integrity, its malfunction can generate detrimental rearrangements of genome or produce toxic intermediates that can cause cell cycle arrest and apoptosis (Fig. 7A [top], B [top]; Fabre et al. 2002). Not surprisingly, cellular mechanisms have evolved that control HR activity, preventing its untimely initiation. Among these mechanisms, disruption of the Rad51-ssDNA filament by displacement of Rad51 from ssDNA seems to be the most efficient one, since it inhibits HR at the early stage (Fig. 7A [top], B [top]). Such a mechanism has been demonstrated for Srs2 helicase from yeast (Krejci et al. 2003; Veaute et al. 2003). Here our results show that BLM helicase can also disrupt the hRad51-ssDNA filament. Thus, BLM may supplement the list of known helicases capable of displacing proteins from nucleic acids in addition to conventional DNA or RNA unwinding activities (Jankowsky et al. 2001; Byrd and Raney 2004).
Although the exact mechanism of hRad51 displacement from ssDNA remains to be investigated, it likely involves BLM translocation along ssDNA, which is consistent with the high ssDNA-dependent ATPase activity of BLM. Another helicase that disrupts the Rad51-ssDNA filament, Srs2, also has strong ssDNA-dependent ATPase activity (Van Komen et al. 2003). In contrast, Rdh54 and Rad54, which displace yeast Rad51 from dsDNA, have dsDNA-dependent ATPase activity and were shown to translocate on dsDNA (Solinger et al. 2002; Amitani et al. 2006; Chi et al. 2006; Holzen et al. 2006). Since BLM physically interacts with hRad51 (Wu et al. 2001), hRad51 displacement may also involve protein–protein interactions. In contrast, BLM does not displace yeast Rad51 from ssDNA, nor does it displace hDmc1, a meiotic hRad51 homolog.
In vivo, the role of the BLM yeast homolog, Sgs1 helicase, in HR has been extensively studied. It was shown that Sgs1 functions overlap with those of Srs2, a helicase that is known to disrupt the Rad51-ssDNA filament (Krejci et al. 2003; Veaute et al. 2003). Thus, overexpression of the Sgs1 helicase can suppress the hyperrecombination phenotype of the srs2Δ strain (Mankouri et al. 2002; Ira et al. 2003). These results are consistent with the role of Sgs1 in disruption of the Rad51-ssDNA filament in vivo.
The ability of BLM to disrupt the hRad51-ssDNA filament depends on the conformation of the filament; BLM can only disrupt an inactive filament in an ADP-bound form. We previously showed that filament inactivation occurs spontaneously during ATP hydrolysis (Bugreev and Mazin 2004). In vitro, an active state of the filament can be preserved in the presence of Ca2+, because Ca2+ specifically inhibits the hydrolytic step of ATP hydrolysis by hRad51. In vivo, some auxiliary proteins may also help to maintain the filament in an active form (Shim et al. 2004). Different sensitivity of the two forms of the filament to BLM disruption suggests a possible mechanism of regulation: Until the cell is fully prepared for HR, the filament remains inactive and susceptible to BLM dissociation. In this case, DNA repair can proceed through different mechanisms; e.g., by nonhomologous end joining, DNA polymerase translesion, or replication fork reversal (Fig. 7A,B).
Our current and previous data (Bugreev et al. 2007) show that the activities of BLM and hRad54 overlap, as both proteins can dissociate D-loops, key intermediates of HR. Moreover, we show that BLM and hRad54 disrupt native D-loops with comparable efficiency. Previously, we suggested that dissociation of D-loops by hRad54, after primer extension by DNA polymerase, may present an important and necessary step that leads to reannealing of two ends of the broken DNA molecule during DSBR through the SDSA mechanism (Bugreev et al. 2007). Sekelsky and coworkers (McVey et al. 2004) suggested a similar function for DmBLM in the SDSA mechanism (Fig. 7C). In contrast, it was also suggested that D-loop dissociation activity of BLM presents a mechanism of suppression of HR (van Brabant et al. 2000; Bachrati et al. 2006). Our current results indicate that these two hypotheses are not necessarily exclusive. The ability of BLM to disrupt “native” D-loops with hRad51 bound to them (which mimic in vivo recombination intermediates) is strongly affected by the conformation of hRad51. Similar to hRad54 protein (Bugreev et al. 2007), BLM cannot dissociate D-loops when hRad51 is present in an active ATP-bound conformation. This property of hRad51 may help to prevent premature disruption of D-loops; i.e., before completion of the primer extension by DNA polymerase. However, if for any reason DNA polymerase is unavailable, BLM may still dissociate D-loops in order to rescue the homologous chromosome that serves as a temple for DSBR (Fig. 7B). In this case, D-loop dissociation would require conversion of the hRad51 filament into an inactive ADP-bound form, as we discussed above.
Sekelsky and coworkers (Adams et al. 2003) found that DmBLM mutants in Drosophila were impaired in their ability to carry out extensive DNA repair synthesis in the SDSA mechanism generating short DNA synthesis tracts. They proposed that BLM can directly stimulate DNA polymerase during DNA repair synthesis by unwinding DNA template ahead of the replication fork. Our current results support this hypothesis, demonstrating that BLM can indeed stimulate DNA polymerase η on DNA substrates that resemble one end of the D-loop, the product of DNA invasion during initial steps of HR. It is interesting to note that such stimulation on genomic DNA, which likely exists in a covalently closed form, would require Topo IIIα and BLAP75, known BLM partners. Topo IIIα and BLAP75 can remove topological constraints from covalently closed substrates, which otherwise would inhibit DNA unwinding activity of BLM (Fig. 7C,D). In yeast, top3 and sgs1 mutants show similar phenotypes, indicating that these two proteins act in concert. Previously it was shown that BLM in a complex with Topo IIIα and BLAP75 promotes resolution of double Holliday junctions in vitro (Fig. 7D; Raynard et al. 2006; Wu et al. 2006). This BLM activity was suggested to suppress crossing over during mitotic recombination. We suggest that BLM may also cooperate with Topo IIIα and BLAP75 promoting DNA repair synthesis in the SDSA mechanism of HR (Fig. 7C,D).
The distribution of functions between D-loop disrupting activities of hRad54 and BLM in the SDSA pathway remains to be investigated. Our data indicate that they may be only partially redundant. Consistent with this view, double RAD54 BLM knockouts in chicken DT-40 cells increase chromosomal instability much above the level observed in each single knockout (Wang et al. 2000). Other activities of BLM and Rad54 may also contribute to the synergistic effect of the double knockouts; e.g., resolution of double Holliday junctions by a complex of BLM with Topo IIIα and BLAP75 or disruption by BLM of the hRad51-ssDNA filament. In addition, our current data demonstrate that only BLM, but not hRad54, can stimulate the primer extension by DNA polymerase η.
In the accompanying paper, Hu et al. (2007) demonstrated that another member of the RecQ family, RECQL5 helicase, can also disrupt the hRad51-ssDNA filament. The available data indicate that BLM and RECQL5 exercise their functions in HR via distinct pathways. Also, whereas RECQL5 seems to specialize in hRad51-ssDNA filament disruption, BLM displays a broad spectrum of activities that either negatively or positively regulate HR at different stages. Taken together, our current results and the results of Hu et al. (2007) establish an important novel function for the RecQ helicase family members: control of HR via the hRad51-ssDNA filament disruption.
Our analysis of the biochemical activities of BLM helicase indicates that this protein may impose a control over HR by either promoting or inhibiting it at different stages (Fig. 7). BLM may prevent premature initiation of recombination events by disrupting the hRad51 filament (Fig. 7A,B). It can also promote HR via the SDSA mechanism by stimulating DNA repair synthesis and catalyzing D-loop dissociation (Fig. 7C). Importantly, the function of BLM may be controlled by other HR proteins; e.g., depending on hRad51 protein conformation, BLM may suppress or promote HR.
Materials and methods
Proteins and DNA
Human Rad51, Rad54, DNA polymerase η, and RPA proteins were purified as described (Henricksen et al. 1994; Masutani et al. 2000; Bugreev and Mazin 2004; Mazina and Mazin 2004). BLM and the BLM K695R mutant were prepared as described (van Brabant et al. 2000) using HisTrap HP and HiTrap Heparin HP columns (GE Healthcare), with an additional chromatographic step on a MonoQ column (GE Healthcare). Supercoiled pUC19 dsDNA was purified as described (Bugreev and Mazin 2004). To prepare linear dsDNA, the pUC19 plasmid was cleaved by SmaI restriction endonuclease (New England Biolabs, Inc.). ϕX174 ssDNA and poly(dT) ssDNA were purchased from New England Biolabs, Inc., and GE Healthcare, respectively. DdeI restriction endonuclease was obtained from New England Biolabs, Inc. All oligonucleotides (IDT, Inc.) used in this study (see Supplementary Table S1) were purified, labeled, and stored as described previously (Bugreev et al. 2006a). Branched DNA, model replication fork, and dsDNA substrates were prepared by the annealing of equimolar amounts of oligonucleotides, as described in Bugreev et al. (2006a).
Inhibition of D-loop formation by BLM
To form the nucleoprotein filament, 32P-labeled ssDNA (#90; 3 μM, nucleotides) was incubated with human or yeast Rad51 protein (1 μM) in buffer containing 25 mM Tris acetate (pH 7.5), 1 mM ATP, 1 mM or 3 mM magnesium acetate (for human and yeast Rad51, respectively), 2 mM DTT, BSA (100 μg/mL), 20 mM phosphocreatine, and creatine phosphokinase (30 U/mL) for 15 min at 37°C. BLM (in indicated concentrations) and hRPA (200 nM) were added, followed by a 10-min incubation. In experiments with hRad51, 2 mM calcium chloride was added, and the reactions were incubated for another 10 min. Then, D-loop formation was initiated by addition of pUC19 scDNA (50 μM, nucleotides). In experiments with yeast Rad51, D-loop formation was initiated by addition of yeast Rad54 (100 nM) and pUC19 scDNA (50 μM, nucleotides). D-loop formation was terminated after a 15-min incubation. The products were deproteinized, analyzed by electrophoresis in a 1% agarose gel, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
D-loop disruption assay
To form D-loops, 32P-labeled tailed DNA (#90; 3 μM, nucleotides) was preincubated with hRad51 protein (1 μM) in buffer containing 25 mM Tris acetate (pH 7.5), 1 mM ATP, 1 mM magnesium acetate, 2 mM calcium chloride, 2 mM DTT, BSA (100 μg/mL), 20 mM phosphocreatine, and creatine phosphokinase (30 U/mL) for 15 min at 37°C. D-loop formation was initiated by addition of hRPA (200 nM) and pUC19 scDNA (50 μM, nucleotides) followed by a 15-min incubation. Then, to remove Ca2+, the reaction mixture was passed through a Bio-Gel P-6 spin column (Bio-Rad) equilibrated with buffer containing 25 mM Tris acetate (pH 7.5), 1 mM ATP, 1 mM magnesium acetate, 2 mM DTT, BSA (100 μg/mL), 20 mM phosphocreatine, and creatine phosphokinase (30 U/mL). Dissociation of “native” D-loops (without removal of hRad51 and hRPA) was initiated by addition of either BLM or hRad54 (in indicated concentrations) and was carried out for 15 min at 37°C. The products of D-loop dissociation were deproteinized, analyzed by electrophoresis in a 1% agarose gel, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Restriction endonuclease protection assay
hRad51-ssDNA nucleoprotein filaments were assembled in reaction buffer containing 25 mM Tris acetate (pH 7.5), 2 mM ATP, 3 mM magnesium acetate, 2 mM DTT, BSA (100 μg/mL), 20 mM phosphocreatine, creatine phosphokinase (30 U/mL), ssDNA (#90; 60 nM, molecules), and hRad51 protein (1 μM) for 15 min at 37°C. BLM (in indicated concentrations) was added, followed by a 5-min incubation. Then, 32P-labeled dsDNA (#25/#26; 15 nM, molecules) was added followed by 10 min incubation. Finally, DdeI restriction endonuclease (0.2 U/μL) was added and, after a 10-min incubation, the DNA products were deproteinized and analyzed by electrophoresis in a 10% polyacrylamide gel. The DNA bands were visualized and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Electron microscopy
hRad51-ssDNA–ATP complexes were formed in 25 mM Triethanolamine-HCl (Fisher) buffer (pH 7.2) with incubations of 15 min at 37°C. hRad51 (6 μM) and M13 ssDNA (Sigma) were present at a ratio of 80:1 (w/w), with ATP (1.5 mM) and magnesium acetate (2 mM). hRad51-hRPA–BLM–ssDNA complexes were formed by adding hRPA (1 μM) and BLM (0.3 μM) to the hRad51-ssDNA–ATP complex solution and were incubated for an additional 15 min at 37°C. hRPA–ssDNA complexes were formed in 25 mM Triethanolamine-HCl (Fisher) buffer (pH 7.2) with incubations of 15 min at 37°C, with hRPA (0.5 μM), and an hRPA to M13 ssDNA ratio of 40:1 (w/w), with magnesium acetate (1 mM). All samples were applied to a carbon film and negatively stained with 2% (w/v) uranyl acetate. The samples were imaged in a Tecnai 12 electron microscope at an accelerating voltage of 80 keV and a magnification on film of 30,000×.
ATPase assay
BLM protein (15 nM) was incubated with indicated DNA cofactors (15 μM, nucleotides) in 10-μL reaction buffer containing 25 mM Tris acetate (pH 7.5), 1 mM ATP, 1 mM magnesium acetate, 25 mM KCl, 2 mM DTT, BSA (100 μg/mL), and 0.1 μCi of [γ-32P]ATP for indicated periods of time at 37°C. The extent of ATP hydrolysis was measured by TLC, as described (Bugreev et al. 2005).
DNA helicase assay
BLM helicase (10 nM) was incubated with a 32P-labeled model replication fork (#185/#193/#363; 10 nM molecules) in DNA polymerase buffer including 70 nM RPA (see below) for 10 min at 37°C. The reactions were terminated and deproteinized by addition of stop buffer up to 1.5% SDS and proteinase K (800 μg/mL) for 15 min at 37°C, mixed with a 0.1 volume of loading buffer (70% glycerol, 0.1% bromophenol blue), and analyzed by electrophoresis in 10% native polyacrylamide gels in TBE buffer at 135 V for 1.5 h To prevent spontaneous reannealing of the DNA products during deproteinization, a 63-mer oligonucleotide (#2; 0.6 μM, molecules) that is complementary to oligonucleotide #193 and nonlabeled oligonucleotide #185 (0.6 μM, molecules) was added with the stop buffer. Gels were dried on DEAE-81 paper (Whatman) and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Stimulation of DNA polymerase η by BLM
A model replication fork was prepared by annealing of 32P-labeled oligonucleotide #185 with oligonucleotides #193 and #363. The reactions containing 10 nM (molecules) 32P-labeled replication fork were carried out in DNA polymerase buffer containing 25 mM Tris acetate (pH 7.5); 2 mM ATP; 7 mM magnesium acetate; 20 mM KCl; 33 μM dATP, dGTP, dCTP, and dTTP; 2 mM DTT; BSA (100 μg/mL); 20 mM phosphocreatine; creatine phosphokinase (30 U/mL); and hRPA (70 nM). BLM helicase or hRad54 were added to the reactions (in indicated concentrations) for 10 min at 37°C. DNA polymerization was started by addition of DNA polymerase η (0.64 nM) followed by a 30-min incubation. The reactions were terminated by ethanol precipitation. The DNA products were dissolved in formamide containing 0.1% bromphenol blue and 0.1% xylene cyanol, heated for 3 min at 95°C, and analyzed by electrophoresis in a 12% denaturing polyacrylamide gel containing 7 M urea.
Acknowledgments
We thank P. Sung, M. Wold, and W. Holloman for hRad51, hRPA, and BLM expression vectors; F. Hanaoka for DNA polymerase η; S. Kowalczykowski for yeast Rad51 and Rad54; R. Brosh Jr. for RecQ1 helicase; and M. Bouchard, M. Rossi, and O. Mazina for the comments and discussion. This work was supported by NIH grants CA100839 to A.V.M. and GM035269 to E.H.E.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1609007
References
- Adams M.D., McVey M., Sekelsky J.J., McVey M., Sekelsky J.J., Sekelsky J.J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Tafel A.A., Kanaar R., Tafel A.A., Kanaar R., Kanaar R. DNA double-strand break repair and chromosome translocations. DNA Repair (Amst.) 2006;5:1075–1081. doi: 10.1016/j.dnarep.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Amitani I., Baskin R.J., Kowalczykowski S.C., Baskin R.J., Kowalczykowski S.C., Kowalczykowski S.C. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D., Hickson I.D. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z., Borts R.H., Hickson I.D., Borts R.H., Hickson I.D., Hickson I.D. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish H., Kedar P., Kaneko H., Chen P., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Kedar P., Kaneko H., Chen P., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Kaneko H., Chen P., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Chen P., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Fukao T., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Peng C., Beresten S., Gueven N., Purdie D., Lees-Miller S., Beresten S., Gueven N., Purdie D., Lees-Miller S., Gueven N., Purdie D., Lees-Miller S., Purdie D., Lees-Miller S., Lees-Miller S., et al. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia-mutated protein, ATM. J. Biol. Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- Bianco P.R., Tracy R.B., Kowalczykowski S.C., Tracy R.B., Kowalczykowski S.C., Kowalczykowski S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Mazin A.V., Mazin A.V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Mazin A.V., Mazin A.V. Reconstitution of dsDNA break repair using human proteins of homologous recombination. Nat. Protoc. 2007 [Google Scholar]
- Bugreev D.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V., Stasiak A.Z., Stasiak A., Mazin A.V., Stasiak A., Mazin A.V., Mazin A.V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Mazina O.M., Mazin A.V., Mazina O.M., Mazin A.V., Mazin A.V. Analysis of branch migration activities of proteins using synthetic DNA substrates. Nat. Protoc. 2006a [Google Scholar]
- Bugreev D.V., Mazina O.M., Mazin A.V., Mazina O.M., Mazin A.V., Mazin A.V. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006b;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Hanaoka F., Mazin A.V., Hanaoka F., Mazin A.V., Mazin A.V. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- Byrd A.K., Raney K.D., Raney K.D. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- Chi P., Kwon Y., Seong C., Epshtein A., Lam I., Sung P., Klein H.L., Kwon Y., Seong C., Epshtein A., Lam I., Sung P., Klein H.L., Seong C., Epshtein A., Lam I., Sung P., Klein H.L., Epshtein A., Lam I., Sung P., Klein H.L., Lam I., Sung P., Klein H.L., Sung P., Klein H.L., Klein H.L. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J. Biol. Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- Cromie G.A., Hyppa R.W., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R., Hyppa R.W., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R., Zakharyevich K., Hunter N., Smith G.R., Hunter N., Smith G.R., Smith G.R. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W.D., Gangloff S., Chan A., Heyer W.D., Gangloff S., Heyer W.D., Gangloff S., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- Gonzalez-Barrera S., Cortes-Ledesma F., Wellinger R.E., Aguilera A., Cortes-Ledesma F., Wellinger R.E., Aguilera A., Wellinger R.E., Aguilera A., Aguilera A. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell. 2003;11:1661–1671. doi: 10.1016/s1097-2765(03)00183-7. [DOI] [PubMed] [Google Scholar]
- Helleday T., Lo J., van Gent D.C., Engelward B.P., Lo J., van Gent D.C., Engelward B.P., van Gent D.C., Engelward B.P., Engelward B.P. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair (Amst.) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht C.B., Wold M.S., Umbricht C.B., Wold M.S., Wold M.S. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Hickson I.D. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Holzen T.M., Shah P.P., Olivares H.A., Bishop D.K., Shah P.P., Olivares H.A., Bishop D.K., Olivares H.A., Bishop D.K., Bishop D.K. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes & Dev. 2006;20:2593–2604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., Stark J.M., Barnes E.L., Chi P., Janscak P., Barnes E.L., Chi P., Janscak P., Chi P., Janscak P., Janscak P., et al. 2007RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments Genes Dev. ,(this issue) 10.1101/gad.1609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N., Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E., Malkova A., Liberi G., Foiani M., Haber J.E., Liberi G., Foiani M., Haber J.E., Foiani M., Haber J.E., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M., Gross C.H., Shuman S., Pyle A.M., Shuman S., Pyle A.M., Pyle A.M. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Chakraverty R.K., Hickson I.D., Chakraverty R.K., Hickson I.D., Hickson I.D. The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Constantinou A., Li J.L., West S.C., Hickson I.D., Constantinou A., Li J.L., West S.C., Hickson I.D., Li J.L., West S.C., Hickson I.D., West S.C., Hickson I.D., Hickson I.D. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Araki K., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Araki K., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Hanaoka F., Nozaki K., Hashimoto N., Nozaki K., Hashimoto N., Hashimoto N., et al. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Reddy M.S., Klein H., Ellenberger T., Sung P., Klein H., Ellenberger T., Sung P., Ellenberger T., Sung P., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Mankouri H.W., Craig T.J., Morgan A., Craig T.J., Morgan A., Morgan A. SGS1 is a multicopy suppressor of srs2: Functional overlap between DNA helicases. Nucleic Acids Res. 2002;30:1103–1113. doi: 10.1093/nar/30.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Iwai S., Hanaoka F., Kusumoto R., Iwai S., Hanaoka F., Iwai S., Hanaoka F., Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina O.M., Mazin A.V., Mazin A.V. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52042–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- Mazina O.M., Rossi M.J., Thomaa N.H., Mazin A.V., Rossi M.J., Thomaa N.H., Mazin A.V., Thomaa N.H., Mazin A.V., Mazin A.V. Interactions of human rad54 protein with branched DNA molecules. J. Biol. Chem. 2007;282:21068–21080. doi: 10.1074/jbc.M701992200. [DOI] [PubMed] [Google Scholar]
- McIlwraith M.J., Vaisman A., Liu Y., Fanning E., Woodgate R., West S.C., Vaisman A., Liu Y., Fanning E., Woodgate R., West S.C., Liu Y., Fanning E., Woodgate R., West S.C., Fanning E., Woodgate R., West S.C., Woodgate R., West S.C., West S.C. Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- McVey M., Larocque J.R., Adams M.D., Sekelsky J.J., Larocque J.R., Adams M.D., Sekelsky J.J., Adams M.D., Sekelsky J.J., Sekelsky J.J. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J.E., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C., Hickson I.D., Wu L., Hickson I.D., Wu L., Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P., Bussen W., Sung P., Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Sharma S., Doherty K.M., Brosh R.M., Doherty K.M., Brosh R.M., Brosh R.M. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K.S., Schmutte C., Tombline G., Heinen C.D., Fishel R., Schmutte C., Tombline G., Heinen C.D., Fishel R., Tombline G., Heinen C.D., Fishel R., Heinen C.D., Fishel R., Fishel R. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J. Biol. Chem. 2004;279:30385–30394. doi: 10.1074/jbc.M306066200. [DOI] [PubMed] [Google Scholar]
- Solinger J.A., Kiianitsa K., Heyer W.D., Kiianitsa K., Heyer W.D., Heyer W.D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Sasaki M.S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S., Sasaki M.S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S., Yamaguchi-Iwai Y., Takata M., Takeda S., Takata M., Takeda S., Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Klein H., Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M.G., Krejci L., Van Komen S., Sehorn M.G., Van Komen S., Sehorn M.G., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- van Brabant A.J., Ye T., Sanz M., German I.J., Ellis N.A., Holloman W.K., Ye T., Sanz M., German I.J., Ellis N.A., Holloman W.K., Sanz M., German I.J., Ellis N.A., Holloman W.K., German I.J., Ellis N.A., Holloman W.K., Ellis N.A., Holloman W.K., Holloman W.K. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Van Komen S., Reddy M.S., Krejci L., Klein H., Sung P., Reddy M.S., Krejci L., Klein H., Sung P., Krejci L., Klein H., Sung P., Klein H., Sung P., Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Kowalczykowski S.C., Le Cam E., Fabre F., Le Cam E., Fabre F., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wang W., Seki M., Narita Y., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T., Seki M., Narita Y., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T., Narita Y., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T., Yamada K., Masuko T., Katada T., Enomoto T., Masuko T., Katada T., Enomoto T., Katada T., Enomoto T., Enomoto T. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Whitby M.C. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D., Hickson I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D., Hickson I.D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies S.L., Levitt N.C., Hickson I.D., Davies S.L., Levitt N.C., Hickson I.D., Levitt N.C., Hickson I.D., Hickson I.D. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Wang W., Li L., Brown G.W., Hickson I.D., Li L., Brown G.W., Hickson I.D., Brown G.W., Hickson I.D., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]