Abstract
Increased methylation of CpG islands and silencing of affected target genes is frequently found in human cancer; however, in vivo the question of causality has only been addressed by loss-of-function studies. To directly evaluate the role and mechanism of de novo methylation in tumor development, we overexpressed the de novo DNA methyltransferases Dnmt3a1 and Dnmt3b1 in ApcMin/+ mice. We found that Dnmt3b1 enhanced the number of colon tumors in ApcMin/+ mice approximately twofold and increased the average size of colonic microadenomas, whereas Dnmt3a1 had no effect. The overexpression of Dnmt3b1 caused loss of imprinting and increased expression of Igf2 as well as methylation and transcriptional silencing of the tumor suppressor genes Sfrp2, Sfrp4, and Sfrp5. Importantly, we found that Dnmt3b1 but not Dnmt3a1 efficiently methylates the same set of genes in tumors and in nontumor tissues, demonstrating that de novo methyltransferases can initiate methylation and silencing of specific genes in phenotypically normal cells. This suggests that DNA methylation patterns in cancer are the result of specific targeting of at least some tumor suppressor genes rather than of random, stochastic methylation followed by clonal selection due to a proliferative advantage caused by tumor suppressor gene silencing.
[Keywords: DNA methylation, epigenetics, cancer, Dnmt3b, APC]
Cancer cells show widespread epigenetic changes when compared with their normal parental tissue, including changes in DNA methylation and chromatin modification (Jones and Baylin 2007). The first epigenetic abnormality reported for human cancer was a global decrease in genomic cytosine methylation (Feinberg and Vogelstein 1983), most often seen in repetitive sequences and intergenic regions. It promotes genetic instability, increases the mobility of transposable elements (Walsh et al. 1998), and induces tumorigenesis in several different mouse models (Eden et al. 2003; Gaudet et al. 2003; Yamada et al. 2005; Jones and Baylin 2007). Thus, hypomethylation predisposes to genetic damage and increases the risk of tumor development. Conversely, in some tissues global hypomethylation can also inhibit tumor outgrowth (Laird et al. 1995).
In addition to global hypomethylation, it was also found that the cancer cell genome frequently contains regions with increased cytosine methylation (Baylin et al. 1986). This regional hypermethylation often affects CpG islands that are associated with promoter regions (Herman and Baylin 2003; Feinberg and Tycko 2004; Jones and Baylin 2007). Regional hypermethylation attracted attention when it was linked to transcriptional silencing of the RB tumor suppressor gene in patients with retinoblastoma tumors (Greger et al. 1989, 1994). Multiple follow-up studies revealed that in cancer many tumor-relevant genes, in particular tumor suppressor genes, are transcriptionally silenced by hypermethylation. Aberrant DNA methylation patterns are stable and maintained throughout cell division, and therefore regional hypermethylation has been proposed to contribute to carcinogenesis (Herman and Baylin 2003). However, when unbiased screens were conducted to identify hypermethylated regions in tumor DNA, many of the affected genes were not directly involved in tumor pathogenesis (Suzuki et al. 2002). These observations stirred a debate about whether the widespread regional hypermethylation in tumors is a consequence rather than a cause of cancer. Two recent discoveries shed more light on the mechanisms and the conceptual understanding of regional hypermethylation in tumors. Firstly, it was found that Polycomb group (PcG) repressor complexes can directly recruit de novo methyltransferases to target genes and promote regional methylation (Vire et al. 2006). Secondly, recent studies demonstrated that hypermethylation in cancer targets developmental regulator genes that are occupied by polycomb repressor complexes in embryonic stem (ES) cells (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). This is consistent with the notion that genes marked by PcG complexes during early development may be particularly susceptible to methylation-mediated silencing.
Epigenetic silencing of genes in cancer poses a number of as yet unresolved questions. For example, an important issue is whether de novo methylation of genes in cancer is causally involved in tumor development, or rather represents an adaptive change reflecting the oncogenic phenotype (Bestor 2003). An early in vitro study using the colon carcinoma cell line HCT116 suggested a causal role of de novo methylation in carcinogenesis (Myohanen et al. 1998). Correlative evidence from human studies is also consistent with regional hypermethylation being causally involved in the tumorigenic process. For example, hypermethylation is already detectable at the earliest stages of tumor development, and some tumor characteristics correlate well with methylation-mediated silencing of certain tumor-relevant genes (Chan et al. 2006). However, most in vivo experiments addressing the role of DNA methylation in cancer are loss-of-function studies using chemical or genetic inhibition of maintenance DNA methylation (Robert et al. 2003) and therefore do not directly address the causal role of hypermethylation in cancer. Such interventions affect global methylation as well as regional methylation, complicating the interpretation of results. One gain-of-function study using in vitro overexpression of Dnmt1 in mammalian cells indeed supported the idea that hypermethylation can promote cell transformation (Wu et al. 1993), but attempts to expand this approach into an adult in vivo model were unsuccessful (Biniszkiewicz et al. 2002). To prove that de novo methylation is causally involved in tumor formation, loss-of-function studies need to be complemented by gain-of-function studies in vivo.
Other open questions relate to the mechanism and specificity of the de novo methylation process: Are altered methylation patterns in tumors the result of sporadic methylation events and selection, or are certain genes directly and efficiently targeted for methylation? Also, the identity and the specificity of DNA-methylating enzymes involved in gene silencing remain unresolved questions. Both de novo DNA methyltransferases (Dnmt) 3a and 3b are frequently expressed in cancer tissue, but their relative contribution to hypermethylation in cancer is unclear. A recent study from our laboratory suggested that Dnmt3b is required for tumor development (Lin et al. 2006), but target genes have not been identified. It remains to be clarified which of the known de novo methyltransferases is involved in tumor-associated hypermethylation in vivo and whether these enzymes have inherent target specificity. To address these questions, we generated several mouse strains that allow for conditional overexpresssion of Dnmt3a1, Dnmt3b1, and isoforms of Dnmt3b1 and studied the effect of these enzymes on tumorigenesis in the ApcMin/+ mouse model for intestinal neoplasia. Both Dnmt3a and Dnmt3b have been shown to mediate de novo methylation of previously unmethylated double-stranded DNA. For example, during embryogenesis, expression of Dnmt3a and Dnmt3b coincides with periods of high de novo methylation activity, and this process is impaired by genetic deletion of these enzymes (Okano et al. 1998, 1999). Both enzymes are therefore considered the major de novo DNA methyltransferases of mammalian cells (Goll and Bestor 2005).
Results
Transgenic mice with inducible Dnmt3a and Dnmt3b transgenes
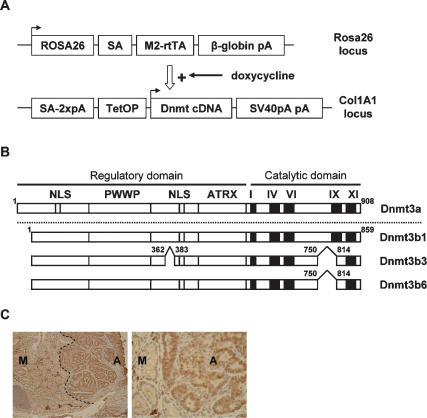
We generated tetracycline-inducible alleles of Dnmt3a1 and Dnmt3b1 and two isoforms of Dnmt3b1—Dnmt3b3 and Dnmt3b6 (Fig. 1A,B)—that are produced by alternative splicing. The isoforms Dnmt3b3 and Dnmt3b6 are truncated versions of the full-length protein Dnmt3b1 and lack part of motif IX of the catalytic domain (Chen et al. 2003). These isoforms are enzymatically inactive and were included in our experiment to allow for a distinction between methylation-related and non-methylation-related effects of Dnmt3b1. The tetracycline-inducible Dnmt3 transgenes were targeted to the 3′ untranslated region (UTR) of the collagen I gene, and the tetracycline-responsive transactivator (M2-rtTA) was inserted into the ROSA26 locus as described previously (Beard et al. 2006). To analyze the effect of Dnmt3a and Dnmt3b activation on tumorigenesis, the respective transgene was crossed into the ApcMin/+ mouse model for intestinal neoplasia, which is heterozygous for the mutant Min allele of the adenomatus polyposis coli (Apc) tumor suppressor gene (Su et al. 1992). Previous experiments from our lab detected increased expression of Dnmt3b in some intestinal adenomas (Fig. 1C) and showed that deletion of Dnmt3b protects against intestinal tumor initiation (Lin et al. 2006). All Dnmt transgenic strains were first backcrossed into the C57BL/6 background and then crossed with ApcMin/+ mice to generate the experimental genotype M2-rtTA+/−, Dnmt transgene+/−, ApcMin/+.
Figure 1.
Transgene constructs. (A) Strategy used to generate transgenic mice with tetracyline-inducible Dnmts: M2-rtTA was targeted to the ROSA26 locus and is under control of the endogenus ROSA26 promotor. An expression cassette containing the respective Dnmt cDNA under control of a tetracycline-responsive promotor was inserted ∼500 base pairs downstream from the 3′ UTR of the collagenA1 gene. (SA) Splice acceptor; (pA) polyadenylation signal; (TetOP) tetracycline-responsive operator. Arrows indicate the transcriptional start site (Beard et al. 2006). (B) Diagram of the Dnmt cDNA constructs used: The position of the conserved PWWP domain, ATRX homology domain, putative NLSs, the methyltransferase motifs (I, IV, VI, and IX), and sites of alternative splicing are shown. Numbers represent amino acids (Chen et al. 2003). (C) Colon section of a control ApcMin/+ mouse, stained with a Dnmt3b antibody (left, 10×; right, 40×). Regions with normal mucosa (M) showed weak expression of Dnmt3b, whereas regions with adenoma tissue (A) showed strong expression of Dnmt3b.
Inducible transgene expression
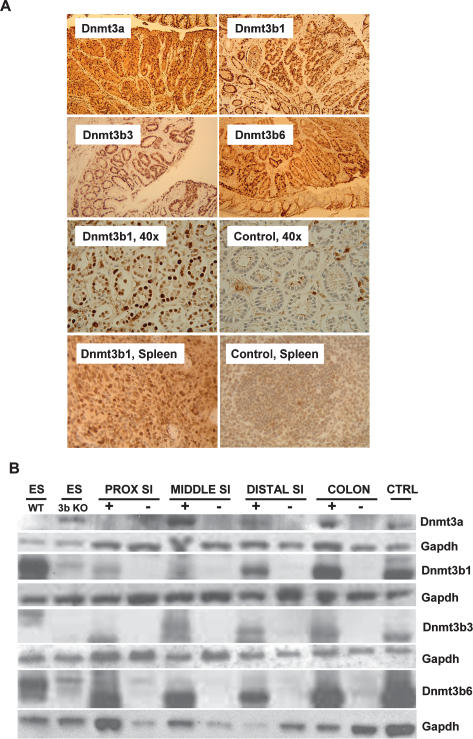
For transgene induction, the experimental animals were fed doxycycline in the drinking water, starting at 4 wk of age, and were compared with the control group that received regular drinking water. The top four panels of Figure 2A show transverse sections of colon samples derived from the four doxycycline-treated mice stained with antibodies against Dnmt3a (top left) or Dnmt3b. Each section covers areas with both normal intestinal mucosa and tumor tissue. The immunostaining demonstrated strong transgene expression in normal epithelial cells of the intestinal mucosa and neoplastic tissue in all experimental groups. The third row of Figure 2A illustrates transgene expression in the colon mucosa at higher magnification from treated (left) and untreated (right) animals. The doxycycline-treated mice showed strong transgene expression with the expected nuclear localization (Fig. 2A), whereas very little or no nuclear staining was detectable in the uninduced mice. Also, Dnmt3b1 was overexpressed in spleen of doxycycline-treated animals (Fig. 2A, bottom row). In general, Dnmt transgene expression was low or undetectable in the nonepithelial cells of the intestinal wall. To compare transgene expression in the different anatomic regions of the intestine, we harvested epithelial cells from the proximal, middle, and distal small intestine and colon from induced and uninduced mice. We found that transgene expression was lower in the proximal small intestine and increased toward the distal intestine, with the highest expression observed in the colon (Fig. 2B; Supplementary Fig. S1). Dnmt3-mediated de novo methylation is thought to require association with the Dnmt3L isoform (Ooi et al. 2007). To assess whether Dnmt3b1 overexpression also induces Dnmt3L, we performed quantitative PCR analyses. Supplementary Figure S2 shows that Dnmt3b1 induction did not alter Dnmt3L expression.
Figure 2.
Transgene expression analysis. (A) Colon and spleen sections of transgenic mice induced with doxycycline and stained with a Dnmt3a (top left) or Dnmt3b antibody. All strains showed clearly visible doxycycline-induced transgene expression in both normal mucosa and adenoma tissue (first and second panel, 10×). The third panel demonstrates appropriate nuclear localization of the transgene product (Dnmt3b1, third panel left, 40×), whereas mucosa from an uninduced control mouse showed weak to no nuclear Dnmt3b staining (third panel, right, 40×). The bottom panel demonstrates that the Dnmt3b1 transgene is also widely expressed in the spleen. (B) Western blot demonstrating doxycycline-inducible protein expression of transgenic mouse strains in different regions of the intestinal mucosa. Top panel is stained with Dnmt3a antibody, and other panels are stained with Dnmt3b antibody; Gapdh was used as loading control. In each case, protein from wild-type ES cells (ES WT) was used as a positive control, and protein from ES cells knocked out for Dnmt3b (ES 3bKO) was used as a negative control. CTRL protein was extracted from immortalized and doxycycline-induced tail tip fibroblasts derived from the same respective mouse strain. All strains showed doxycycline-inducible transgene expression. In general, Dnmt expression was lowest in the proximal small intestine (SI) and highest in the colon. This is particularly the case with Dnmt3b1. The weak band in the ES 3bKO lane (runnning slightly above the Dnmt3b1 band), which is seen in blots with Dnmt3b antibody, is most likely nonspecific.
Dnmt3b1 activation increases the number of intestinal adenomas
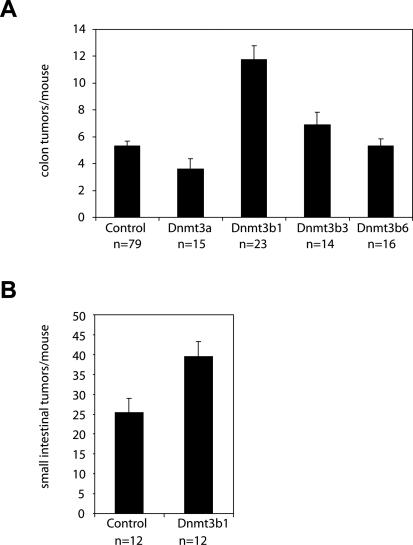
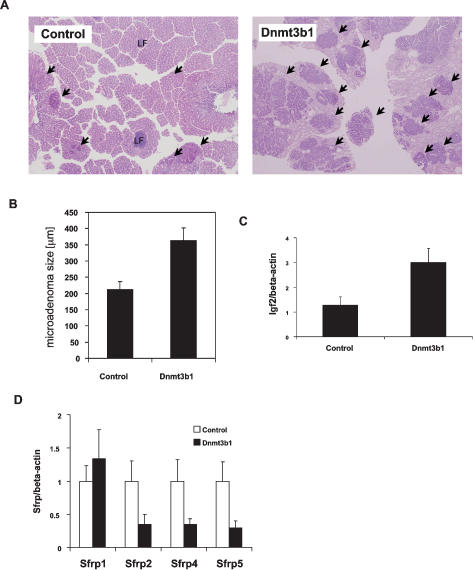
When analyzing the colon of experimental mice at 5 mo of age, we found a more than twofold increase in macroscopic tumors in transgenic mice expressing full-length Dnmt3b1 when compared with controls (11.7 ± 1 vs. 5.3 ± 0.3 colon tumors/mouse; n = 23 and n = 79, respectively; P < 0.0002, Mann-Whitney U-test, values given as mean ± SE), and this difference was statistically significant. All other transgenic strains showed either very small or no changes in colon tumor numbers when compared with controls (Fig. 3A). Expression of full-length Dnmt3b1 also significantly increased the number of small intestinal adenomas (39.5 ± 3.7 vs. 25.3 ± 3.6 small intestinal tumors/mouse; n = 12 each, P < 0.01, Mann-Whitney U-test, values given as mean ± SE), with the relative increase being less pronounced than in the colon, possibly due to lower expression of the transgene (Fig. 3B). Tumor formation in the intestine occurs in two stages: microscopic adenomas and macroscopic adenomas, where microadenomas are considered precursor lesions to macroscopic tumors (Yamada et al. 2002; Lin et al. 2006; Yamada and Mori 2007). To evaluate whether Dnmt3b1 overexpression affects these early tumor stages, we analyzed the morphology of microadenomas in the colon of Dnmt3b1-expressing mice using horizontal en face sections of the colon mucosa. Figure 4, A and B shows that the average size of microadenomas was increased almost twofold in mice expressing Dnmt3b1 when compared with controls (364 μm ± 38 vs. 211 μm ± 25; n = 58 and n = 70, respectively, P < 0.0005, Mann-Whitney U-test, values given as mean ± SE). No difference was found in the ratio of microscopic to macroscopic adenomas when comparing control mice with Dnmt3b1-expressing mice (ratio microdaenomas/macroadenomas control: 3.6 ± 1.9; Dnmt3b1: 4.8 ± 1.6, n = 5 each, P = 0.75). These findings suggest that Dnmt3b1 overexpression affects the earliest stages of tumor development.
Figure 3.
Dnmt3b1 overexpression increases the number of intestinal tumors. (A) Number of macroscopic colon tumors per mouse. Increased expression of Dnmt3b1 caused a 2.2-fold increase in the number of colon tumors per mouse (11.7 ± 1 vs. 5.3 ± 0.3 colon tumors per mouse; P < 0.0002, Mann-Whitney test). Other mouse strains showed small changes (Dnmt3b3, 1.3-fold increase, P < 0.02; Dnmt3a, 0.68-fold decrease, P < 0.01) or no change (Dnmt3b6) in colon tumor numbers. (B) Number of small intestinal adenomas per mouse. Expression of Dnmt3b1 caused a 1.6-fold increase in small intestinal adenomas (39.5 ± 3.7 vs. 25.3 ± 25.3 small intestinal tumors per mouse, P < 0.02, Mann-Whitney test). Values are given as mean ± SE.
Figure 4.
Dnmt3b1 overexpression increases the size of colonic microadenomas, increases expression of Igf2, and decreases expression of Sfrp2, Sfrp4, and Sfrp5 in colon tumors. (A) Horizontal en face sections of colon mucosa derived from a control and Dnmt3b1-expressing mouse (H&E stain, 10×). (Arrows) Microadenomas; (LF) lymph follicule. The size of microadenomas in the mouse with increased Dnmt3b1 expression was much larger than in the control mouse. (B) The average size of colonic microadenomas in Dnmt3b1-expressing mice was increased 1.7-fold when compared with controls (364 μm ± 38 vs. 211 μm ± 25 colon; P < 0.0005). Values represent mean ± SE. (C) Quantitation of Igf2 expression in colon tumors normalized to β-actin expression using real-time PCR. The relative Igf2 expression in tumors derived from Dnmt3b1 mice (2.9 ± 0.6, n = 10) was significantly higher than Igf2 expression in control mice (1.3 ± 0.4, n = 11), P < 0.03 Mann Whitney U-test. (D) Quantitation of Sfrp expression in colon tumors normalized to β-actin expression using real-time PCR. To facilitate comparisons in one graph, measurements of each Sfrp RNA were normalized to the average value of the respective control group. The expression of Sfrp2, Sfrp4, and Sfrp5 was significantly lower in tumors derived from Dnmt3b1 mice (Sfrp2, P < 0.002; control, 1 ± 0.3, n = 10; Dnmt3b1, 0.35 ± 0.2, n = 10; Sfrp4, P < 0.05; control, 1 ± 0.3, n = 9; Dnmt3b1: 0.26 ± 0.15, n = 10; Sfrp5, P < 0.003; control, 1 ± 0.3, n = 9; Dnmt3b1: 0.29 ± 0.1, n = 10, Mann-Whitney U-test). Values represent mean ± SE.
Dnmt3b1 promotes loss of imprinting of Igf2
The increased tumorigenesis by Dnmt3b1 overexpression but not by enzymatically inactive isoforms is consistent with silencing of genes involved in tumor formation by hypermethylation. To identify possible target genes, we analyzed genes that are known to be silenced in cancer by de novo methylation. Insulin-like growth factor II (Igf2) has been shown to promote intestinal tumorigenesis in mouse models, and increased Igf2 expression in humans due to loss of imprinting has been linked to an increased risk of colon cancer (Cui et al. 2003). To analyze whether overexpression of Dnmt3b1 caused loss of imprinting and increased expression of Igf2, we first compared the Igf2 expression in colon tumors from control mice with Igf2 expression in tumors derived from Dnmt3b1-overexpressing mice using quantitative PCR. Figure 4C shows that Igf2 expression was increased almost twofold in tumors from Dnmt3b1-expressing mice when compared with tumors from control mice. Bisulfite sequencing analysis of the H19 DMR showed that tumors from Dnmt3b1-expressing mice were hypermethylated at this locus, when compared with controls (Fig. 5A). Analysis of the non-tumor-bearing mucosa also showed a slight increase in H19 DMR methylation in Dnmt3b1-expressing mice, but to a lesser degree than in the macroscopic tumors (Fig. 5A). Both the expression and the bisulfite sequencing data are consistent with biallelic expression of Igf2 due to loss of imprinting. We used liquid chromatography coupled to a mass spectrometer (LC–MS/MS) to assess whether Dnmt3b1 overexpression altered the overall DNA methylation level. Supplementary Figure S3 shows that the 5-methylcytosine content of DNA from Dnmt3b1-overexpressing mice did not differ from control samples, suggesting that global DNA methylation was not affected by Dnmt3b1 overexpression.
Figure 5.
Dnmt3b1 overexpression causes biallelic methylation of the H19 DMR but does not affect methylation of the Snrpn DMR1. (A) Bisulfite sequencing analysis of the H19 DMR of tumor DNA and colon mucosa DNA derived from control (right columns) and Dnmt3b1-expressing (left columns) mice (five tumors each and three colon mucosa samples each). The region analyzed covers 16 CpG sites. Each horizontal line represents one individual sequence; each block of sequences represents data from one sample. (Black boxes) Methylated CpGs; (white boxes) unmethylated CpGs. For each sample, the biallelic or monoallelic status of the methylation pattern was assumed from the ratio of methylated and unmethylated alleles. Four out of five Dnmt3b1-expressing tumors showed clearly increased H19 DMR methylation, whereas all control tumors showed mostly monoallelic methylation. (B) Bisulfite sequencing analysis of the Snrpn DMR1 of tumor DNA and colon mucosa DNA derived from control (right column) and Dnmt3b1-expressing (left column) mice (five tumors each and three colon mucosa samples each). Sixteen CpG sites were analyzed. Only one Dnmt3b1-expressing tumor showed clear hypermethylation at this site. A second tumor was possibly hypermethylated; however, the number of sequences is low. All colon mucosa samples show monoallelic methylation.
Methylation of the Snurf/Snrpn imprinting center
To evaluate whether Dnmt3b1 expression also affects imprinted regions that do not promote cell growth and are therefore not subject to positive selection, we analyzed methylation of the differentially methylated region 1 (DMR1) of the Snurf/Snrpn imprinting center. This imprinting center is normally methylated on the suppressed maternal allele, with the unmethylated paternal allele being expressed. Methylation of this imprinting center negatively controls expression of the Snurf/Snrpn genes, which are predominantly expressed in the developing and adult brain (Reinhart et al. 2006). Bisulfite sequencing analysis indeed showed that in contrast to the H19 DMR, the monoallelic methylation pattern of this region was not significantly altered in Dnmt3b1-overexpressing mice (Fig. 5B).
Binding, methylation, and transcriptional down-regulation of Sfrp family members by Dnmt3b1
Activation of the Wnt pathway is a key component of most intestinal tumors (Clevers 2006). We therefore investigated whether Dnmt3b1 expression contributed to epigenetic silencing of secreted frizzled-related protein (Sfrp) genes, which are considered Wnt pathway inhibitors (Suzuki et al. 2004). We first analyzed the expression of Sfrp1, frp2, Sfrp4, and Sfrp5 in colon tumors from experimental and control mice using quantitative PCR. Figure 4D shows that expression of Sfrp2, Sfrp4, and Sfrp5 was significantly down-regulated in tumors derived from Dnmt3b1-expressing mice when compared with controls. Based on this finding, we conducted bisulfite sequencing analysis of the promoter regions of Sfrp2, Sfrp4, and Sfrp5. Figure 6A shows that methylation of Sfrp2 and 5 genes was significantly increased in Dnmt3b1-expressing tumors, when compared with controls. Methylation of Sfrp4 was also increased, as shown in Supplementary Figure S4. Importantly, analysis of the tumor-free mucosa also showed a significant increase in Sfrp2 and Sfrp5 methylation in samples derived from Dnmt3b1-overexpressing mice, and for Sfrp2 this finding was confirmed in Apc wild-type mice (Fig. 6B). This suggests that overexpression of Dnmt3b1 directly targets Sfrp2, Sfrp4, and Sfrp5 for de novo methylation and silencing without clonal selection of cells carrying randomly silenced tumor suppressor genes being involved. In contrast to Dnmt3b1, Dnmt3a1 overexpression did not cause significant methylation of Sfrp2 (Fig. 6A). Sfrp2 was not de novo methylated in spleen (Supplementary Fig. S5), suggesting tissue specificity of gene silencing.
Figure 6.
Dnmt3b1 causes hypermethylation of Sfrp2 and Sfrp5 in both tumors and normal colon epithelial cells. (A) Bisulfite sequencing analysis of the Sfrp2 and Sfrp5 promoter region of tumor DNA derived from control mice, Dnmt3b1-expressing mice, and Dnmt3a1-expressing mice (Sfrp2). In each case, five colon tumors from five different mice were analyzed. In the case of Sfrp2, 14 CpG sites were analyzed, and in the case of Sfrp5, 20 CpG sites were analyzed. Four out of five Dnmt3b1-expressing tumors showed clearly increased methylation at the Sfrp2 promoter, and all five such tumors showed increased methylation at the Sfrp5 promoter, when compared with control tumors. No significant methylation of Sfrp2 was detected in tumors derived from Dnmt3a1-overexpressing mice. (B) Bisulfite sequencing analysis of the Sfrp2 and Sfrp5 promoter region of DNA derived from non-tumor-bearing colon epithelial cells (colon mucosa). Sfrp2 methylation was analyzed in four Apc heterozygote mice (+/−) and in three Apc wild-type mice with Dnmt3b1 overexpression. Both Apc wild-type mice and Apc heterozygote mice show a clear increase in Sfrp2 methylation following Dnmt3b1 expression when compared with controls, suggesting that Dnmt3b1 directly methylates this target in the absence of coexisting pathology. (Bottom right) Methylation of Sfrp5 is also increased in colon epithelial cells from Dnmt3b1-expressing mice when compared with controls.
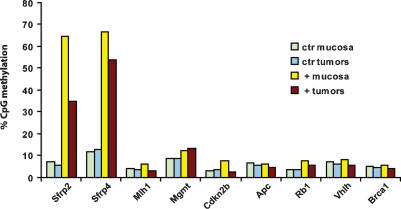
To further evaluate the target specificity of Dnmt3b1, we analyzed the methylation pattern of three CpG islands associated with the Cdx2 promoter region. The tumor suppressor gene Cdx2 is transcriptionally down-regulated in tumors from ApcMin/+ mice and therefore is a potential target for de novo methylation. However, in contrast to the Sfrp genes, we found that the Cdx2-associated CpG islands were completely unmethylated in Dnmt3b1-overexpressing tumors (Supplementary Fig. S6), supporting the hypothesis that Dnmt3b1 targets specific sequences. To screen for additional tumor-relevant targets of Dnmt3b1, we also analyzed the methylation status of Mlh1, Mgmt, Cdkn2b, Apc, Rb1, Vhlh, and Brca1 by MALDI-TOF mass spectrometry, using the previously identified targets Sfrp2 and Sfrp4 as positive controls. Figure 7 demonstrates that none of these additional loci was methylated by Dnmt3b1, whereas methylation of Sfrp2 and 4 was confirmed, supporting the concept that this enzyme targets only specific loci. We also analyzed whether Dnmt3b1 was directly bound to methylated target loci, using chromatin immunoprecipitation (ChIP). The results (Supplementary Fig. S7) show that the Dnmt3b1 protein was physically associated with the DMRs of the methylated target loci Sfrp2, Sfrp4, Sfrp5, and H19 but not with the control loci Cdx2 and β-actin, consistent with Dnmt3b1 binding to selected target genes.
Figure 7.
Dnmt3b1 does not cause hypermethylation of the tumor relevant genes Mlh1, Mgmt, Cdkn2b, Apc, Rb1, Vhlh, and Brca1. The methylation status of seven tumor relevant target genes (Mlh1, Mgmt, Cdkn2b, Apc, Rb1, Vhlh, and Brca1) was analyzed by MALDI-TOF mass spectrometry, using two previously identified methylation targets (Sfrp2 and Sfrp4) as positive controls. We analyzed DNA from colon epithelial cells and colon tumors derived from Dnmt3b1-overexpressing mice and uninduced control mice (Dnmt3b1+ colon, n = 4; control colon, n = 3; Dnmt3b1+ tumors, n = 5; control tumors, n = 4). The average CpG methylation rate was calculated for each amplicon (one amplicon per gene) of each experimental group, and these averages are shown in the bar graph. The data demonstrate that none of these seven genes show signs of increased methylation upon overexpression of Dnmt3b1, whereas methylation of Sfrp2 and Sfrp4 is confirmed, further suggesting that Dnmt3b1 only methylates specific genetic loci.
Apc loss of heterozygosity (LOH) analysis
The ApcMin/+ mouse strain contains one wild-type (+) and one mutant (Min) allele of the Apc tumor suppressor. Loss of function of Apc is an important component of tumor development in these mice, and LOH of this gene has been detected in the majority of intestinal tumors. We therefore investigated whether epigenetic inactivation of the Apc wild-type allele (+) might have contributed to the increased tumor load found in Dnmt3b1-expressing mice. We isolated DNA from colon tumors with Dnmt3b1 overexpression and from control tumors and analyzed the frequency of Apc LOH. Supplementary Figure S8 shows that in both groups almost all tumors showed loss of the wild-type Apc allele. This indicates that epigenetic loss of Apc function is unlikely to contribute to the increased tumor load of Dnmt3b1-expressing mice and supports the methylation analysis of the Apc locus, which suggested that this gene was not methylated by Dnmt3b1 (Fig. 7). Only one tumor in the Dnmt3b1 group appeared to have retained the wild-type allele. It is possible that in this particular tumor aberrant methylation contributed to silencing of the wild-type allele and loss of Apc function; however, the low frequency of this event cannot account for the observed increase in tumor numbers.
Discussion
Our study focused on three main goals: to evaluate a possible cause–effect relationship between de novo methylation and tumor development, to establish an experimental system that can unmask and help identify genes susceptible to de novo methylation, and to analyze how different de novo methyltransferases affect regional DNA methylation in vivo.
To address these questions, we generated four transgenic mouse strains that allow for conditional overexpression of full-length Dnmt3a1, full-length Dnmt3b1, and two isoforms of Dnmt3b1 and studied the effect of overexpression on tumor development in the ApcMin/+ model. We found that full-length Dnmt3b1 increased the number of intestinal adenomas and microadenomas, whereas the truncated isoforms Dnmt3b3 and Dnmt3b6 had a small, if any, effect on the tumor number. Both Dnmt3b3 and Dnmt3b6 have no enzymatic activity because they lack part of the methyltransferase catalytic domain (Chen et al. 2002, 2003). It is therefore likely that the observed tumor-promoting effect of Dnmt3b1 is mediated by DNA methylation. This distinction is important because it had been shown that Dnmt3b can suppress transcription in the absence of DNA-methylating activity (Bachman et al. 2001). When evaluating possible molecular targets of Dnmt3b1-mediated methylation, we followed a candidate approach. We selected genes for expression and methylation analyses that are relevant for intestinal tumor development and are known targets for de novo methylation in cancer.
Igf2 is an imprinted gene encoding a single-chain polypetide with paracrine and autocrine activity that promotes mitosis mostly through signaling via the Igf-1 receptor (Dupont et al. 2003). Igf2 has been shown to promote tumor development in mouse models (Hassan and Howell 2000; Sakatani et al. 2005), and its increased expression in humans correlates with an increased risk for tumor development, in particular colorectal cancer (Cui et al. 2003; Woodson et al. 2004). Monoallelic expression of Igf2 depends on the methylation status of the imprinting control region located between the Igf2 and the H19 gene (H19 DMR). The unmethylated maternal allele allows binding of the CTCF insulator protein, which blocks downstream enhancer sequences from interacting with the upstream Igf2 gene. Methylation of this sequence inhibits binding of the insulator protein and allows expression of the respective Igf2 allele. Thus, de novo methylation of the H19 DMR results in loss of imprinting and increased expression of Igf2 (Bell and Felsenfeld 2000; Hark et al. 2000; Kurukuti et al. 2006). In a recent study, it was demonstrated that loss of imprinting and biallelic expression of Igf2, due to deletion of the H19 DMR on the maternal allele, causes a twofold increase in intestinal tumors in ApcMin/+ mice (Sakatani et al. 2005), which is similar to the relative tumor increase that we observed. This suggests that the H19 DMR hypermethylation and increased Igf2 expression are partially responsible for the increased tumor load found in Dnmt3b1-expressing mice. Enhanced Igf2 expression is also a possible explanation for the increased size of microscopic adenomas observed in these animals. Our observations provide direct in vivo evidence that loss of imprinting of Igf2 can be caused by de novo methylation, as opposed to genetic rearrangements that cause loss of the maternal and duplication of the paternal Igf2 allele (Haddad and Held 1997; Cooper et al. 2007). Deletion of Dnmt3b did not affect size or number of microadenomas, suggesting that the phenotype of the Dnmt3b1-overexpressing mice is not complementary to the phenotype of the Dnmt3b knockout model (Lin et al. 2006).
To evaluate whether Dnmt3b1 expression results in general loss of imprinting, we analyzed the DMR1 of the Snurf/Snrpn imprinting center (Reinhart et al. 2006). Snurf/Snrpn is a bicistronic imprinted gene encoding two polypeptides: the SmN splicing factor, which is involved in RNA processing, and the Snurf polypeptide of unknown function. Snrpn also encodes a long (∼460-kb) alternatively spliced RNA transcript that contains several families of snoRNAs. A direct effect on cell proliferation has not been reported for the products of this locus, suggesting that growth-mediated selection of methylation events at this imprinting center is unlikely (Rodriguez-Jato et al. 2005). In contrast to the H19 DMR, we found that most samples from Dnmt3b1-expressing mice maintained normal imprinting patterns at this site. This suggests that the H19 DMR is either inherently more susceptible to de novo methylation by Dnmt3b1 or that cells with loss of imprinting of Igf2 have a selective growth advantage.
In addition to Igf2, we also analyzed the expression and methylation status of Sfrp genes. Five members of this protein family have been identified (Sfrp1–5). They contain regions homologous to the frizzled receptor, including a cysteine-rich domain, which allows for binding and sequestering of Wnt ligands. These proteins are therefore considered inhibitors of the Wnt pathway (Jones and Jomary 2002). Importantly, it has been shown that the Sfrp proteins 1, 2, 4, and 5 inhibit the activity of the Wnt pathway even in the presence of downstream activating mutations, such as loss of function of Apc (Suzuki et al. 2004). Each of these genes is associated with a CpG island that meets the formal criteria published by Takai and Jones (2002). We found that Sfrp2, Sfrp4, and Sfrp5 are significantly down-regulated in tumors derived from Dnmt3b1 mice and that the promoter regions of Sfrp2, Sfrp4, and Sfrp5 are significantly de novo methylated in tumor samples from Dnmt3b1 mice when compared with controls. Importantly, increased methylation of Sfrp2 and Sfrp5 was already detectable in the normal intestinal mucosa. Since Sfrp proteins can inhibit the Wnt pathway, and methylation and down-regulation of Sfrp genes is strongly associated with colorectal cancer in human patients (Caldwell et al. 2004; Suzuki et al. 2004), these observations suggest that epigenetic silencing of these targets contributed to the increased tumor load of Dnmt3b1-expressing mice. Our results show that de novo DNA methylation can induce silencing of Sfrp genes, supporting the view that DNA methylation is a cause rather than a consequence of transcriptional silencing (Gu et al. 2006). This concept is also supported by the observation that the Cdx2 gene remained unmethylated in Dnmt3b1-overexpressing tumors, even though it is transcriptionally down-regulated in ApcMin/+ tumors. To further evaluate the specificity of de novo methylation by Dnmt3b1, we analyzed the methylation status of seven additional tumor-relevant genes, selecting candidates from different functional categories, such as DNA repair (Mlh1, Mgmt, and Brca1), Cdk inhibitor (Cdkn2b), and tumor suppressor genes (Apc, Rb1, and Vhlh). None of these loci was hypermethylated upon Dnmt3b1 overexpression, supporting the hypothesis that Dnmt3b1 targets and methylates specific genetic loci.
Little is known about the relative role of different de novo methyltransferases in cancer. In contrast to Dnmt3b1, Dnmt3a1 overexpression did not cause any significant methylation of Sfrp2, which is consistent with the observation that Dnmt3a1 overexpression did not enhance tumor development. Several studies have shown different roles for Dnmt3a and Dnmt3b in development. For example, Dnmt3a is required for methylation of imprinted loci during gametogenesis, whereas Dnmt3b is not (Kaneda et al. 2004). Also, major satellite repeats of the pericentromeric region are preferentially methylated by Dnmt3a, whereas minor satellite repeats are methylated by Dnmt3b (Chen et al. 2003). A recent study identified a specific gene (Fgf-1) that is preferentially methylated by Dnmt3a and not by Dnmt3b (Oka et al. 2006). Our findings suggest that these differences may be significant for tumor development and that Dnmt3b1 is possibly a more relevant mediator of de novo methylation in cancer than Dnmt3a.
Our analysis of Apc LOH in colon tumors shows that the majority of tumors derived from Dnmt3b1-expressing mice lost the Apc wild-type allele, which is similar to control mice. It is therefore unlikely that epigenetic silencing of the Apc wild-type allele contributed to the increased tumor load of Dnmt3b1-overexpressing mice. Hypermethylation of the APC gene has been reported for selected cases of intestinal tumors in FAP patients, where loss of APC function was documented and at the same time LOH of the APC gene was excluded (Esteller et al. 2000, 2001). However, even though hypermethylation of the APC gene was found in these selected cases, it was not detected in unbiased screens to identify methylation targets in tumors, and in our own study we also did not detect any methylation of this locus (Weber et al. 2005; Keshet et al. 2006; Widschwendter et al. 2007).
An important question is whether the epigenetic changes induced by Dnmt3b1 overexpression correspond to stochastic epimutations with subsequent selection due to proliferative advantage or whether the enzyme specifically targets some genes more efficiently than others. Our observations suggest that the aberrant DNA methylation in our model is localized and not stochastic. First, analysis of Dnmt3b1-overexpressing tissue showed no increase in global 5-methylcytosine content when compared with control tissue, supporting the idea that this DNA methyltransferase affects regional methylation and not global methylation (Supplementary Fig. S3). Secondly, we found that overexpression of one de novo methyltransferase (Dnmt3b1) causes hypermethylation of Sfrp2, whereas overexpression of another de novo methyltransferase (Dnmt3a1) does not, suggesting differential targeting of these enzymes. Third, and more importantly, methylation of affected target regions by Dnmt3b1 was detectable not only in clonal tumor tissue but also in normal colon mucosa. Because in normal mucosa selection for tumor suppressor gene silencing is unlikely, our results support the hypothesis that Dnmt3b1 can cause sequence-specific de novo methylation of some tumor suppressor genes. This concept is further supported by the methylation screen of seven additional tumor-relevant target loci, which did not show de novo methylation in mucosa or tumor samples from Dnmt3b1-overexpressing mice. In contrast to the normal intestinal mucosa, Sfrp2 was not methylated in the spleen, even though the Dnmt3b1 transgene was expressed in this tissue. This suggests that the specificity of Dnmt3b1-mediated de novo methylation may be influenced by tissue-specific chromatin states of target genes.
The results of our study raise several interesting questions with regard to the mechanisms that mediate specific targeting of Dnmt3b1 to certain loci. Is targeting specificity an inherent property of Dnmt3b1, or are additional binding partners such as Dnmt3L required for recruitment? What role does the DNA sequence play, and which chromatin states of target genes are required for this process? Reports on the DNA sequence specificity of methylation targets in cancer are conflicting. One study suggested enrichment of certain sequence motifs in tumor-related methylation targets (Keshet et al. 2006), whereas another study could not detect any sequence specificity of de novo methylation in cancer (Weber et al. 2005). In contrast to DNA sequence specificity, recent studies found good correlations between the chromatin state of genes and their methylation frequency in cancer; in particular, H3K27 marks correlated with an increased frequency of DNA methylation in tumors (Schlesinger et al. 2007; Widschwendter et al. 2007). Sfrp2, Sfrp4, and Sfrp5 are reported targets of polycomb complexes and are marked by H3K27 methylation in mouse and human ES cells (Boyer et al. 2006; Lee et al. 2006a). Similarly, when we analyzed the expression of Runx3, which is also a PcG target and in humans is frequently methylated in colon cancer (Widschwendter et al. 2007), we found reduced expression in tumors derived from Dnmt3b1-overexpressing mice (data not shown). On the other hand, we could not detect any methylation of the Cdx2 gene, which is occupied by PcG complexes in ES cells (Boyer et al. 2006; Lee et al. 2006a), and the H19 DMR, which was methylated in our study, reportedly does not contain any H3K27 marks (Mikkelsen et al. 2007). Clearly, methylation analysis of more regions combined with histone mapping is necessary to allow a more detailed correlation of Dnmt3b1-mediated de novo methylation and histone modifications.
At this point, it is unclear whether binding of Dnmt3b1 to specific target sites is an inherent property of the protein itself, or if additional mediators such as Dnmt3L are required for this process (Ooi et al. 2007). However, Dnmt3L expression was not increased upon overexpression of Dnmt3b1. Initial ChIP experiments suggested that Dnmt3b1 was indeed physically bound to the same loci that were found to be methylated (Supplementary Fig. S7), consistent with its role in de novo methylation of specific target genes. Our results support recent models on the contribution of epigenetic changes to tumor development: Based on the finding that epigenetic suppression of tumor relevant genes is often already detectable in preneoplastic tissue, it has been proposed that regional hypermethylation predisposes these tissues to later tumor development (Baylin and Ohm 2006; Feinberg et al. 2006; Jones and Baylin 2007). In our model, Dnmt3b1 expression establishes a methylation pattern that is already detectable in non-tumor-bearing tissues. This is consistent with the hypothesis that epigenetic rather than genetic alterations may be the very first changes in the process that converts a normal to a malignant cell (Baylin and Ohm 2006; Feinberg et al. 2006; Jones and Baylin 2007). Further analyses will also help clarify whether inappropriate activation of Dnmt3b is the molecular basis for the CpG island methylator phenotype (CIMP), which is found in a subset of human cancers (Weisenberger et al. 2006).
Materials and methods
DNA constructs and targeting of ES cells
Plasmids containing the coding region of the respective Dnmts were kindly provided by Dr. En Li (Novartis Institute of Medicine, Cambridge, MA). Briefly, the respective Dnmt cDNA was cloned into the HindIII–EcoRI sites (for Dnmt3a1) or the BamHI–EcoRI sites (for Dnmt3b1 and isoforms) of the recipient plasmid pcDNA6/V5-HisA (Invitrogen) (Chen et al. 2002). In our laboratory, these coding regions were released from pcDNA6/V5-HisA using HindIII/EcoRI (for Dnmt3a1) and BamHI/EcoRI (for Dnmt3b1 and isoforms) and inserted into the EcoRI site of the targeting vector pBS31 via blunt-end ligation. All final targeting vectors were verified by sequencing before transfection into ES cells. Targeting of transgenes to the ColA1 locus of KH2 cells was conducted as described previously (Beard et al. 2006). Properties of the targeting vector are detailed in the Supplemental Material. Clones with correct integration of the targeting vector into the homing site of KH2 cells were identified by hygromycin resistance and Southern blotting.
Generation of mice, genotyping, and transgene induction
Transgenic ES cells were used to generate mice as described previously (Beard et al. 2006). Mice with tet-inducible Dnmt alleles were backcrossed to the C57BL/6 strain for four generations prior to crossing with ApcMin/+ mice. C57BL/6 ApcMin/+ mice were obtained from The Jackson Laboratory. All final experimental mice analyzed in this study had been backcrossed for at least six generations to the C57BL/6 background. For genotyping, DNA from tail tips was PCR-amplified using primers and conditions listed in the Supplemental Material. For transgene induction, mice were fed 0.5 mg/mL doxycycline in the drinking water supplemented with 10 mg/mL sucrose. Control mice received regular drinking water.
Tissue harvesting and tumor analysis
All experimental mice were sacrificed at 5 mo of age, and macroscopic and microscopic adenomas were analyzed as described previously (Yamada et al. 2005). Counting was conducted blind with regard to genotype. P values were obtained using the Mann-Whitney U-test. Colon epithelial cells were harvested from tumor-free colon samples as described previously (Fujimoto et al. 2002).
Histological analysis, immunostaining, Western blotting, and ChIP
Paraffin sections (5-μm) were used for hematoxylin and eosin (H&E) staining and immunohistochemical analysis. Primary antibodies for immunostaining were anti-Dnmt3a (monoclonal, 1:150 dilution, Imgenex) and anti-Dnmt3b (monoclonal, 1:150 dilution, Imgenex). For Western blotting, we used anti-Dnmt3a (chicken polyclonal, 1:2000 dilution, Abcam), anti-Dnmt3b (monoclonal, 1:500 dilution, Imgenex), and anti-GAPDH (rabbit polyclonal, 1:2000, Abcam). ChIP was conducted as described previously (Lee et al. 2006b), using the Dnmt3b antibody ab2851 (rabbit polyclonal, Abcam). Primer pairs used for quantitative PCR following ChIP are listed in the Supplemental Material.
cDNA and real-time PCR
cDNA was generated from total RNA using oligo-dT primers. For each sample, a minus-RT reaction was generated, and pooled minus-RT reactions were used as negative controls for quantitative PCR reactions (SYBR Green, Invitrogen). For quantitation, standard dilution curves were included on each plate. All samples were analyzed in triplicate; β-actin was used as an endogenous control. Individual data were converted to relative values based on the standard curve and then normalized to the β-actin values of the same sample. Primers used are listed in the Supplemental Material.
Bisulfite sequencing, methylation screen, 5-methylcytosine quantitation, and Apc LOH analysis
For bisulfite sequencing analysis, DNA was mutagenized using the Epitect kit (Qiagen). Target regions were PCR-amplified and subcloned, and minipreps of individual colonies were sequenced. Only sequences with >95% non-CpG C conversion were used for analysis. Primers and PCR conditions for individual loci are listed in the Supplemental Material. Methylation screening using MALDI-TOF MS was conducted as described previously (Ehrich et al. 2005). Primer pairs used for this analysis are listed in the Supplemental Material. Quantitative analysis of 5-methylcytosine content of DNA was performed by LC–MS/MS as described previously (Crain 1990). 2′-deoxycytidine (dC), and 5-methyl-2′-deoxycytidine (5mdC) were monitored in the analysis, and 2′-deoxycytidine (15N3) was used as an internal standard. LOH of the Apc gene was analyzed by PCR as described previously (Yamada et al. 2002).
Acknowledgments
We thank Jessica Dausman, Ruth Flannery, and Dongdong Fu for help with maintenance of the mouse colony and histological analysis. We are also very grateful for the help of Tom DiCesare in the preparation of figures. Heinz Linhart is supported by a post-doctoral fellowship from the Fritz-Thyssen Stiftung. This work was supported by a research grant from Philip Morris Products S.A. and National Institutes of Health grant NIH CA087869 to R.J.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1594007
References
- Bachman K.E., Rountree M.R., Baylin S.B., Rountree M.R., Baylin S.B., Baylin S.B. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Ohm J.E., Ohm J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Hoppener J.W., de Bustros A., Steenbergh P.H., Lips C.J., Nelkin B.D., Hoppener J.W., de Bustros A., Steenbergh P.H., Lips C.J., Nelkin B.D., de Bustros A., Steenbergh P.H., Lips C.J., Nelkin B.D., Steenbergh P.H., Lips C.J., Nelkin B.D., Lips C.J., Nelkin B.D., Nelkin B.D. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–2922. [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R., Hochedlinger K., Plath K., Wutz A., Jaenisch R., Plath K., Wutz A., Jaenisch R., Wutz A., Jaenisch R., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Bell A.C., Felsenfeld G., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. Unanswered questions about the role of promoter methylation in carcinogenesis. Ann. N. Y. Acad. Sci. 2003;983:22–27. doi: 10.1111/j.1749-6632.2003.tb05959.x. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Jun Z., Walter J., Jaenisch R., Walter J., Jaenisch R., Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Levine S.S., Wernig M., Tajonar A., Ray M.K., Wernig M., Tajonar A., Ray M.K., Tajonar A., Ray M.K., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Caldwell G.M., Jones C., Gensberg K., Jan S., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Jones C., Gensberg K., Jan S., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Gensberg K., Jan S., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Jan S., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Chughtai S., Wallis Y., Matthews G.M., Morton D.G., Wallis Y., Matthews G.M., Morton D.G., Matthews G.M., Morton D.G., Morton D.G. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–888. doi: 10.1158/0008-5472.can-03-1346. [DOI] [PubMed] [Google Scholar]
- Chan T.L., Yuen S.T., Kong C.K., Chan Y.W., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Yuen S.T., Kong C.K., Chan Y.W., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Kong C.K., Chan Y.W., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Chan Y.W., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., Lo M.W., Tam W.Y., Li V.S., Tam W.Y., Li V.S., Li V.S., et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat. Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Chen T., Ueda Y., Xie S., Li E., Ueda Y., Xie S., Li E., Xie S., Li E., Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- Chen T., Ueda Y., Dodge J.E., Wang Z., Li E., Ueda Y., Dodge J.E., Wang Z., Li E., Dodge J.E., Wang Z., Li E., Wang Z., Li E., Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cooper W.N., Curley R., Macdonald F., Maher E.R., Curley R., Macdonald F., Maher E.R., Macdonald F., Maher E.R., Maher E.R. Mitotic recombination and uniparental disomy in Beckwith-Wiedemann syndrome. Genomics. 2007;89:613–617. doi: 10.1016/j.ygeno.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Crain P.F. Analysis of 5-methylcytosine in DNA by isotope dilution gas chromatography–mass spectrometry. Methods Enzymol. 1990;193:857–865. doi: 10.1016/0076-6879(90)93455-t. [DOI] [PubMed] [Google Scholar]
- Cui H., Cruz-Correa M., Giardiello F.M., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Cruz-Correa M., Giardiello F.M., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Giardiello F.M., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P., Wu Y., He X., Powe N.R., Feinberg A.P., He X., Powe N.R., Feinberg A.P., Powe N.R., Feinberg A.P., Feinberg A.P. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Dupont J., Pierre A., Froment P., Moreau C., Pierre A., Froment P., Moreau C., Froment P., Moreau C., Moreau C. The insulin-like growth factor axis in cell cycle progression. Horm. Metab. Res. 2003;35:740–750. doi: 10.1055/s-2004-814162. [DOI] [PubMed] [Google Scholar]
- Eden A., Gaudet F., Waghmare A., Jaenisch R., Gaudet F., Waghmare A., Jaenisch R., Waghmare A., Jaenisch R., Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Xinarianos G., Cantor C.R., Field J.K., van den Boom D., Cantor C.R., Field J.K., van den Boom D., Field J.K., van den Boom D., van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M., Sparks A., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Sparks A., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., Tarafa G., Sidransky D., Meltzer S.J., Sidransky D., Meltzer S.J., Meltzer S.J., et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- Esteller M., Fraga M.F., Guo M., Garcia-Foncillas J., Hedenfalk I., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Fraga M.F., Guo M., Garcia-Foncillas J., Hedenfalk I., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Guo M., Garcia-Foncillas J., Hedenfalk I., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Garcia-Foncillas J., Hedenfalk I., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Hedenfalk I., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Godwin A.K., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Trojan J., Vaurs-Barriere C., Bignon Y.J., Ramus S., Vaurs-Barriere C., Bignon Y.J., Ramus S., Bignon Y.J., Ramus S., Ramus S., et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum. Mol. Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Tycko B., Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Vogelstein B., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Ohlsson R., Henikoff S., Ohlsson R., Henikoff S., Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Fujimoto K., Beauchamp R.D., Whitehead R.H., Beauchamp R.D., Whitehead R.H., Whitehead R.H. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- Gaudet F., Hodgson J.G., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R., Hodgson J.G., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R., Dausman J., Gray J.W., Leonhardt H., Jaenisch R., Gray J.W., Leonhardt H., Jaenisch R., Leonhardt H., Jaenisch R., Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Greger V., Passarge E., Hopping W., Messmer E., Horsthemke B., Passarge E., Hopping W., Messmer E., Horsthemke B., Hopping W., Messmer E., Horsthemke B., Messmer E., Horsthemke B., Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- Greger V., Debus N., Lohmann D., Hopping W., Passarge E., Horsthemke B., Debus N., Lohmann D., Hopping W., Passarge E., Horsthemke B., Lohmann D., Hopping W., Passarge E., Horsthemke B., Hopping W., Passarge E., Horsthemke B., Passarge E., Horsthemke B., Horsthemke B. Frequency and parental origin of hypermethylated RB1 alleles in retinoblastoma. Hum. Genet. 1994;94:491–496. doi: 10.1007/BF00211013. [DOI] [PubMed] [Google Scholar]
- Gu P., Le Menuet D., Chung A.C., Cooney A.J., Le Menuet D., Chung A.C., Cooney A.J., Chung A.C., Cooney A.J., Cooney A.J. Differential recruitment of methylated CpG binding domains by the orphan receptor GCNF initiates the repression and silencing of Oct4 expression. Mol. Cell. Biol. 2006;26:9471–9483. doi: 10.1128/MCB.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Haddad R., Held W.A., Held W.A. Genomic imprinting and Igf2 influence liver tumorigenesis and loss of heterozygosity in SV40 T antigen transgenic mice. Cancer Res. 1997;57:4615–4623. [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M., Ingram R.S., Levorse J.M., Tilghman S.M., Levorse J.M., Tilghman S.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hassan A.B., Howell J.A., Howell J.A. Insulin-like growth factor II supply modifies growth of intestinal adenoma in ApcMin/+ mice. Cancer Res. 2000;60:1070–1076. [PubMed] [Google Scholar]
- Herman J.G., Baylin S.B., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.E., Jomary C., Jomary C. Secreted Frizzled-related proteins: Searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Sado T., Tsujimoto N., Li E., Sasaki H., Tsujimoto N., Li E., Sasaki H., Li E., Sasaki H., Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Rand E., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Hecht M., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Segal E., Pikarski E., Young R.A., Niveleau A., Cedar H., Pikarski E., Young R.A., Niveleau A., Cedar H., Young R.A., Niveleau A., Cedar H., Niveleau A., Cedar H., Cedar H., et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Zhao Z., Lobanenkov V., Reik W., Ohlsson R., Lobanenkov V., Reik W., Ohlsson R., Reik W., Ohlsson R., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P.W., Jackson-Grusby L., Fazeli A., Dickinson S.L., Jung W.E., Li E., Weinberg R.A., Jaenisch R., Jackson-Grusby L., Fazeli A., Dickinson S.L., Jung W.E., Li E., Weinberg R.A., Jaenisch R., Fazeli A., Dickinson S.L., Jung W.E., Li E., Weinberg R.A., Jaenisch R., Dickinson S.L., Jung W.E., Li E., Weinberg R.A., Jaenisch R., Jung W.E., Li E., Weinberg R.A., Jaenisch R., Li E., Weinberg R.A., Jaenisch R., Weinberg R.A., Jaenisch R., Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Johnstone S.E., Cole M.F., Isono K., Cole M.F., Isono K., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006a;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., Young R.A., Johnstone S.E., Young R.A., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006b;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Yamada Y., Nguyen S., Linhart H., Jackson-Grusby L., Meissner A., Meletis K., Lo G., Jaenisch R., Yamada Y., Nguyen S., Linhart H., Jackson-Grusby L., Meissner A., Meletis K., Lo G., Jaenisch R., Nguyen S., Linhart H., Jackson-Grusby L., Meissner A., Meletis K., Lo G., Jaenisch R., Linhart H., Jackson-Grusby L., Meissner A., Meletis K., Lo G., Jaenisch R., Jackson-Grusby L., Meissner A., Meletis K., Lo G., Jaenisch R., Meissner A., Meletis K., Lo G., Jaenisch R., Meletis K., Lo G., Jaenisch R., Lo G., Jaenisch R., Jaenisch R. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol. Cell. Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Alvarez P., Brockman W., Kim T.K., Koche R.P., Brockman W., Kim T.K., Koche R.P., Kim T.K., Koche R.P., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myohanen S.K., Baylin S.B., Herman J.G., Baylin S.B., Herman J.G., Herman J.G. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- Ohm J.E., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., Mohammad H.P., Chen W., Daniel V.C., Yu W., Chen W., Daniel V.C., Yu W., Daniel V.C., Yu W., Yu W., et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka M., Rodic N., Graddy J., Chang L.J., Terada N., Rodic N., Graddy J., Chang L.J., Terada N., Graddy J., Chang L.J., Terada N., Chang L.J., Terada N., Terada N. CpG sites preferentially methylated by Dnmt3a in vivo. J. Biol. Chem. 2006;281:9901–9908. doi: 10.1074/jbc.M511100200. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie S., Li E., Xie S., Li E., Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E., Bell D.W., Haber D.A., Li E., Haber D.A., Li E., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Tempst P., Lin S.P., Allis C.D., Lin S.P., Allis C.D., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B., Paoloni-Giacobino A., Chaillet J.R., Paoloni-Giacobino A., Chaillet J.R., Chaillet J.R. Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol. Cell. Biol. 2006;26:8347–8356. doi: 10.1128/MCB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert M.F., Morin S., Beaulieu N., Gauthier F., Chute I.C., Barsalou A., MacLeod A.R., Morin S., Beaulieu N., Gauthier F., Chute I.C., Barsalou A., MacLeod A.R., Beaulieu N., Gauthier F., Chute I.C., Barsalou A., MacLeod A.R., Gauthier F., Chute I.C., Barsalou A., MacLeod A.R., Chute I.C., Barsalou A., MacLeod A.R., Barsalou A., MacLeod A.R., MacLeod A.R. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Jato S., Nicholls R.D., Driscoll D.J., Yang T.P., Nicholls R.D., Driscoll D.J., Yang T.P., Driscoll D.J., Yang T.P., Yang T.P. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatani T., Kaneda A., Iacobuzio-Donahue C.A., Carter M.G., de Boom Witzel S., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Kaneda A., Iacobuzio-Donahue C.A., Carter M.G., de Boom Witzel S., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Iacobuzio-Donahue C.A., Carter M.G., de Boom Witzel S., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Carter M.G., de Boom Witzel S., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., de Boom Witzel S., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Okano H., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Ko M.S., Ohlsson R., Longo D.L., Feinberg A.P., Ohlsson R., Longo D.L., Feinberg A.P., Longo D.L., Feinberg A.P., Feinberg A.P. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., Ben-Shushan E., Reubinoff B.E., Reubinoff B.E., et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Su L.K., Kinzler K.W., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., Dove W.F., Kinzler K.W., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., Dove W.F., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., Dove W.F., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., Dove W.F., Moser A.R., Luongo C., Gould K.A., Dove W.F., Luongo C., Gould K.A., Dove W.F., Gould K.A., Dove W.F., Dove W.F. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Gabrielson E., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Chen W., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Anbazhagan R., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., van Engeland M., Weijenberg M.P., Herman J.G., Baylin S.B., Weijenberg M.P., Herman J.G., Baylin S.B., Herman J.G., Baylin S.B., Baylin S.B. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Watkins D.N., Jair K.W., Schuebel K.E., Markowitz S.D., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Watkins D.N., Jair K.W., Schuebel K.E., Markowitz S.D., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Jair K.W., Schuebel K.E., Markowitz S.D., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Schuebel K.E., Markowitz S.D., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Markowitz S.D., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Chen W.D., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Pretlow T.P., Yang B., Akiyama Y., Van Engeland M., Yang B., Akiyama Y., Van Engeland M., Akiyama Y., Van Engeland M., Van Engeland M., et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Takai D., Jones P.A., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Van Eynde A., Bernard D., Vanderwinden J.M., Bernard D., Vanderwinden J.M., Vanderwinden J.M., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H., Chaillet J.R., Bestor T.H., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Haase M., Lam W.L., Schubeler D., Lam W.L., Schubeler D., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Kang G.H., Widschwendter M., Weener D., Buchanan D., Widschwendter M., Weener D., Buchanan D., Weener D., Buchanan D., Buchanan D., et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., Weisenberger D.J., Campan M., Young J., Jacobs I., Campan M., Young J., Jacobs I., Young J., Jacobs I., Jacobs I., et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Woodson K., Flood A., Green L., Tangrea J.A., Hanson J., Cash B., Schatzkin A., Schoenfeld P., Flood A., Green L., Tangrea J.A., Hanson J., Cash B., Schatzkin A., Schoenfeld P., Green L., Tangrea J.A., Hanson J., Cash B., Schatzkin A., Schoenfeld P., Tangrea J.A., Hanson J., Cash B., Schatzkin A., Schoenfeld P., Hanson J., Cash B., Schatzkin A., Schoenfeld P., Cash B., Schatzkin A., Schoenfeld P., Schatzkin A., Schoenfeld P., Schoenfeld P. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J. Natl. Cancer Inst. 2004;96:407–410. doi: 10.1093/jnci/djh042. [DOI] [PubMed] [Google Scholar]
- Wu J., Issa J.P., Herman J., Bassett D.E., Nelkin B.D., Baylin S.B., Issa J.P., Herman J., Bassett D.E., Nelkin B.D., Baylin S.B., Herman J., Bassett D.E., Nelkin B.D., Baylin S.B., Bassett D.E., Nelkin B.D., Baylin S.B., Nelkin B.D., Baylin S.B., Baylin S.B. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Mori H., Mori H. Multistep carcinogenesis of the colon in ApcMin/+ mouse. Cancer Sci. 2007;98:6–10. doi: 10.1111/j.1349-7006.2006.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Hata K., Hirose Y., Hara A., Sugie S., Kuno T., Yoshimi N., Tanaka T., Mori H., Hata K., Hirose Y., Hara A., Sugie S., Kuno T., Yoshimi N., Tanaka T., Mori H., Hirose Y., Hara A., Sugie S., Kuno T., Yoshimi N., Tanaka T., Mori H., Hara A., Sugie S., Kuno T., Yoshimi N., Tanaka T., Mori H., Sugie S., Kuno T., Yoshimi N., Tanaka T., Mori H., Kuno T., Yoshimi N., Tanaka T., Mori H., Yoshimi N., Tanaka T., Mori H., Tanaka T., Mori H., Mori H. Microadenomatous lesions involving loss of Apc heterozygosity in the colon of adult ApcMin/+ mice. Cancer Res. 2002;62:6367–6370. [PubMed] [Google Scholar]
- Yamada Y., Jackson-Grusby L., Linhart H., Meissner A., Eden A., Lin H., Jaenisch R., Jackson-Grusby L., Linhart H., Meissner A., Eden A., Lin H., Jaenisch R., Linhart H., Meissner A., Eden A., Lin H., Jaenisch R., Meissner A., Eden A., Lin H., Jaenisch R., Eden A., Lin H., Jaenisch R., Lin H., Jaenisch R., Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc. Natl. Acad. Sci. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]