Abstract
Small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), are essential regulatory molecules of many cellular processes. Arabidopsis has at least three classes of endogenous siRNAs—chromatin-associated siRNAs, trans-acting siRNAs (tasiRNAs), and natural antisense transcript (NAT)-associated siRNAs (nat-siRNAs)—all 20–25 nucleotides (nt) in length. Here, we identified a novel class of small RNAs, long siRNAs (lsiRNAs), which are 30–40 nt and share many common features with known siRNAs. The lsiRNAs identified so far are induced by pathogen infection or under specific growth conditions. One of the lsiRNAs, AtlsiRNA-1, is generated from SRRLK/AtRAP NAT pair and specifically induced by the bacterium Pseudomonas syringae carrying effector avrRpt2. Recently, 25- to 31-nt PIWI-interacting RNAs (piRNAs) and repeat-associated siRNAs (rasiRNAs) were identified in animal germline cells. In contrast to the biogenesis of piRNAs/rasiRNAs, which is dicer independent and requires PIWI subfamily proteins, generation of AtlsiRNA-1 requires DCL1, DCL4, and the ARGONAUTE subfamily protein AGO7. It also depends on HYL1, HEN1, HST1, RDR6, and Pol IV. Induction of AtlsiRNA-1 silences AtRAP, which encodes a RAP-domain protein involved in disease resistance. Our further analysis implies that AtlsiRNA-1 may destabilize target mRNA through decapping and XRN4-mediated 5′-to-3′ degradation.
[Keywords: Long siRNAs (lsiRNAs), DCL1, AGO7, decapping, bacteria-induced]
Small RNA-mediated gene silencing plays important roles in many cellular processes, including development, genome maintenance and integrity, and adaptive responses to biotic and abiotic stress in most eukaryotes. Small RNAs, usually 20–25 nucleotides (nt) in length, can be grouped into two classes on the basis of their origins: microRNAs (miRNAs) and small interfering RNAs (siRNAs). They guide heterochromatin formation, mRNA degradation, translational inhibition, and DNA elimination (Baulcombe 2004; Zamore and Haley 2005; Vazquez 2006). Rapid and phenomenal progress has been achieved in unraveling the components and mechanisms involved in the biogenesis and function of various small RNAs. In plants, small RNAs are highly diverse. In general, miRNAs in Arabidopsis are processed from single-stranded (ss) hairpin RNA precursors by an RNase III-type enzyme Dicer-like (DCL) 1. A recent study shows that a couple of newly identified miRNAs are processed by DCL4 (Rajagopalan et al. 2006). The precursors of these miRNAs tend to have more complementarity within the foldback structure than that in most previously identified DCL1-dependent miRNAs. miRNAs are primarily associated with ARGONAUTE 1 (AGO1) to guide mRNA cleavage or translational inhibition. Endogenous siRNAs are usually processed from long double-stranded RNAs (dsRNAs). The generation of trans-acting siRNAs (tasiRNAs) is initiated by miRNAs, and requires DCL4 and RNA-dependent RNA polymerase (RDR) 6 for their biogenesis. Both miRNAs and tasiRNAs are 21- to 22-nt. The most abundant siRNA class is the 24- to 25-nt chromatin-associated siRNAs, which are generated by a DCL3/RDR2/RNA polymerase (Pol) IV-dependent pathway and mainly incorporated into AGO4 to induce transcriptional silencing. Another class of endogenous siRNAs, nat-siRNAs, is derived from natural antisense transcript (NAT) regions (Borsani et al. 2005; Wang et al. 2005; Katiyar-Agarwal et al. 2006; Henz et al. 2007). These are 21- to 24-nt RNA molecules generated by DCL1 and/or DCL2 (Borsani et al. 2005; Katiyar-Agarwal et al. 2006).
Recently, the finding of a novel group of small RNAs with a distinctly larger size, 24–31 nt, added a new dimension in small RNA biology. The 27- to 30-nt small scan RNAs (scnRNAs) were identified in a ciliate protozoan, Tetrahymena thermophila, to direct developmentally programmed DNA elimination (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004; Lee and Collins 2006). PIWI-interacting RNAs (piRNAs) of 26- to 31-nt were discovered in mammalian germline cells (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006) and zebrafish (Houwing et al. 2007). Drosophila repeat-associated siRNAs (rasiRNAs) are 24- to 29-nt, and direct silencing of retrotransposons and repetitive sequences to stabilize the genome (Aravin et al. 2001, 2003; Vagin et al. 2006). rasiRNAs share many common features with piRNAs and are also specifically associated with PIWI proteins (Saito et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007). The accumulation of Tetrahymena scnRNAs requires the Dicer-like protein DCL1P (Mochizuki and Gorovsky 2005), whereas the biogenesis of rasiRNAs in Drosophila appears to be independent of Dicer-1 or Dicer-2 (Vagin et al. 2006). The 5′-end formation of the piRNAs/rasiRNAs involves the slicer activity of the interacting PIWI subfamily proteins (Brennecke et al. 2007; Gunawardane et al. 2007). The accumulation of Drosophila rasiRNA requires two putative nucleases: Zucchini and Squash (Pane et al. 2007). Whether these nucleases are responsible for the formation of rasiRNA 3′ ends needs to be experimentally determined. In plants, two small RNAs (30 and 24 nt), complementary to the locus 3′ to the canonical poly(A) site of FLOWERING LOCUS (FLC), were recently detected in Arabidopsis (Swiezewski et al. 2007). However, the function and biogenesis of the 30-nt small RNA has not been explored.
Here, we describe the identification of a new class of 30- to 40-nt endogenous small RNAs in Arabidopsis. These small RNAs share many common features with other known endogenous siRNAs, therefore, we named them long siRNAs (lsiRNAs). The lsiRNAs identified so far are induced either in response to pathogen infection or under specific growth conditions. One of the lsiRNAs, AtlsiRNA-1, is specifically and strongly induced in response to Pseudomonas syringae pv. tomato (Pst) carrying the effector avrRpt2. We further deciphered the components required for the biogenesis of AtlsiRNA-1, DCL1, DCL4, HEN1, HYL1, HST1, AGO7, RDR6, and Pol IV, which are distinct from those involved in biogenesis of miRNAs and other siRNAs in plants reported so far. Our genetic data suggest that the induction of AtlsiRNA-1 leads to down-regulation of its target, AtRAP mRNA, possibly by promoting mRNA decapping and 5′-to-3′ degradation. The knockout mutant of AtRAP shows enhanced resistance to both virulent Pst and avirulent Pst (avrRpt2) infection.
Results
Long noncoding small RNA species exist in Arabidopsis
Some miRNAs and endogenous siRNAs are induced by pathogen infection and may play a regulatory role in plant immunity (Katiyar-Agarwal et al. 2006; Navarro et al. 2006). To identify more small RNAs specifically induced in response to pathogen attack, we performed in silico analysis of small RNAs that target pathogen-regulated genes from the AtGenExpress data (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp; H. Jin, V. Vacic, T. Girke, S. Lonardi, and J.K. Zhu, in prep.) using publicly available small RNA databases—the Arabidopsis Small RNA Project (ASRP) (Gustafson et al. 2005) and Massively Parallel Signature Sequencing (MPSS) (Lu et al. 2005), and a data set recently published by David Bartel and coworkers (Rajagopalan et al. 2006). Since current small RNA datasets were generated mainly from nonstress-treated material (Llave et al. 2002; Gustafson et al. 2005; Lu et al. 2005; Rajagopalan et al. 2006; Fahlgren et al. 2007; Kasschau et al. 2007), we also performed a pilot small RNA profiling experiment on biotic stress-challenged plants using 454 sequencing (http://www.454.com; Margulies et al. 2005). The biotic stress conditions included bacterial infection by virulent Pst and avirulent Pst (avrRpt2) strains and fungal infection by Alternaria brassicicola or Botrytis cinerea (H. Jin, V. Vacic, T. Girke, S. Lonardi, and J.K. Zhu, in prep.). During the validation of a list of small RNAs by Northern blot analysis, we identified several novel small RNAs ranging from 30-nt to 40-nt (Fig. 1). We named this new class of small RNAs long siRNAs (AtlsiRNAs). The AtlsiRNA-1 signature was detected in the MPSS database and derived from At2g31880 (Supplementary Table S1). AtlsiRNA-2, AtlsiRNA-3, and AtlsiRNA-4 were detected in our 454 sequencing biotic stress data set and corresponded to At4g19680, At4g02970, and At4g00360, respectively (Fig. 1A–D; Supplementary Table S1). AtlsiRNA-1 and AtlsiRNA-4 are derived from the NAT regions. In an effort to identify possible nat- siRNAs from selected NAT pairs using strand-specific hydrolyzed probes spanning the overlapping regions, we detected two more lsiRNAs, AtlsiRNA-5 and AtlsiRNA-6, derived from the overlapping regions of At4g01690/At4g01700 and At5g58120/At5g58130, respectively (Fig. 1E,F).
Figure 1.
Expression of lsiRNAs in Arabidopsis. (A–E) Expression of AtlsiRNA1–5 at 15 hpi of Pst DC3000 carrying an empty vector (EV) or the effector gene avrRpt2, avrRpm1, avrRps4, or avrPphB. AtlsiRNA-2 is also detected in the leaves sprayed with 2 × 105 spores per milliliter Alternaria brassicicola and harvested 2 and 4 dpi. (F) Expression of AtlsiRNA-6 in cell suspension culture. A diagram of genome position for each lsiRNA (black arrowhead) is shown below each panel. The arrowheads above the gene indicate the lsiRNAs with the same sequence as the gene, whereas the arrowheads below the gene indicate the lsiRNAs with the complementary sequence to the gene. Seventy-five micrograms of low-molecular-weight (LMW) RNA were used in each sample. The oligonucleotide probes complementary to the lsiRNA sequences were used for A–D (Supplementary Table S1). Ribo-probes synthesized for the overlapping region complementary to the sense genes At4g01700 and At5g58120 are employed for E and F. (The bars show the probe region.) U6 RNA was used as a control for measuring the relative abundance (RA) (shown below). Ethidium bromide-stained tRNA is shown as a loading control.
These lsiRNAs are mainly induced by bacterial infection or specific developmental growth conditions. For example, AtlsiRNA-1 to AtlsiRNA-5 were induced in response to Pst (avrRpt2) infection (Fig. 1A–E); AtlsiRNA-6 was specifically expressed in cell suspension culture and cannot be detected in seedlings or leaves (Fig. 1F; data not shown). In contrast to animal piRNAs, which are primarily generated from retroelements and repetitive loci and are specific to germline cells, these lsiRNAs are generated from protein-coding genes and are expressed in various tissues or cell types, including mature leaves, seedlings, or cell culture (Fig. 1; data not shown).
To address the biogenesis and functional relevance of this new class of small RNAs in plants, we performed a case study of AtlsiRNA-1 because it was specifically induced by Pst (avrRpt2) infection and showed the strongest signal in our Northern analysis (Fig. 1A). This small RNA was derived from the overlapping region of a NAT pair, At2g31880/At2g31890. At2g31880 encodes a putative leucine-rich repeat receptor-like protein kinase (RLK), and At2g31890 encodes an expressed protein that contains a putative RNA-binding domain, RAP (RNA-binding domain abundant in Apicomplexans) domain. We named these two genes small RNA-generating RLK (SRRLK) and AtRAP, respectively. Since MPSS provided only 17-nt sequences, to obtain more sequence information, we constructed a 20- to 50-nt small-RNA library from avrRpt2-infected leaves harvested at 14 h post-inoculation (hpi). Five distinct 3′ ends and one more distinct 5′ end, besides the 5′ end sequenced by MPSS, were detected by 3′ and 5′ rapid amplification of cDNA ends (RACE) with 24-nt gene-specific primers based on the MPSS sequence and adaptor-specific primers (Supplementary Fig. S1A). These data indicate that AtlsiRNA-1 is a cluster of heterogeneous lsiRNAs, most being 39–41 nt (based on Northern blot analysis) (Fig. 1A; Supplementary Fig. S1A). This heterogeneous phenomenon was observed at many endogenous small RNA loci, even at many miRNA loci in plants (ASRP database http://asrp.cgrb.oregonstate.edu; Rajagopalan et al. 2006).
To eliminate the possibility that AtlsiRNA-1 could be a byproduct of a miRNA, we predicted the secondary RNA structure of this region using Mfold (http://bioweb.pasteur.fr). AtlsiRNA-1 is present in the stem-like region, where no other miRNAs could be predicted (Supplementary Fig. S1B). We also experimentally checked for the presence of additional small RNAs arising from the overlap region by using oligonucleotide probes (probes 1–6) complementary to the flanking regions of AtlsiRNA-1 (Supplementary Fig. S2). No clear signal was detected in untreated or Pst (avrRpt2)-treated samples. Furthermore, we employed three strand-specific ribo probes complementary to the entire overlapping region but excluding the AtlsiRNA-1 site and detected no clear signal (data not shown). These data suggest that no additional small RNAs are induced to a detectable level from the overlapping region of SRRLK and AtRAP in response to Pst (avrRpt2) infection. The accumulation of small RNAs complementary to AtlsiRNA-1 could hardly be detected (probe 7) (Supplementary Fig. S2).
Biogenesis of AtlsiRNA-1 involves components of distinct small RNA pathways
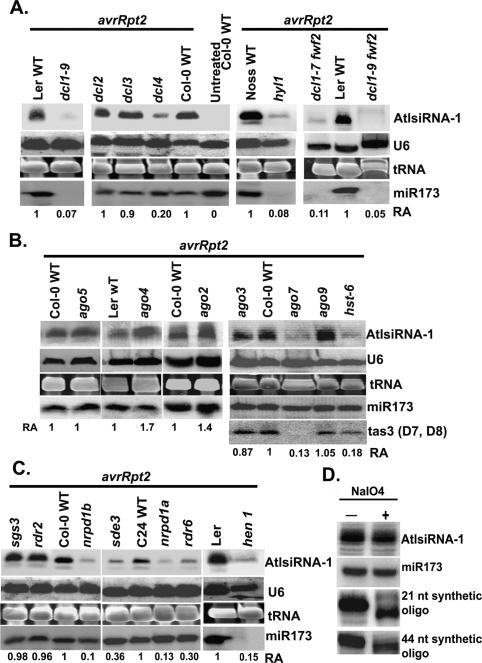
The next important question was what components are involved in the generation of AtlsiRNA-1. Although all the miRNAs and siRNAs studied so far require Dicer or Dicer-like proteins for their biogenesis, piRNAs/ rasiRNA biogenesis is Dicer independent but requires PIWI proteins. Arabidopsis contains four Dicer-like proteins and 10 AGO proteins but no PIWI subclass proteins. To determine the components required for AtlsiRNA-1 formation, we challenged different small-RNA biogenesis mutants and their corresponding background ecotypes with Pst (avrRpt2) and examined the expression of AtlsiRNA-1 at 14 hpi. The level of AtlsiRNA-1 in dcl3-1 and dcl2-1 mutants was similar to that in the wild-type control but significantly reduced in dcl1-9 as compared with the Ler wild type (Fig. 2A). To rule out the marked reduction of AtlsiRNA-1 in dcl1-9 being a secondary effect of its strong morphological phenotype, we tested double homozygous dcl1-7 fwf-2 and dcl1-9 fwf-2 mutants, which have rescued pleiotropic phenotypes and viability (A. Vivian-Smith and R. Offringa, in prep.). The level of AtlsiRNA-1 was also greatly reduced in these double mutants, which confirmed the requirement of DCL1 in AtlsiRNA-1 formation and showed its independence of dcl1-9 morphology (Fig. 2A). AtlsiRNA-1 was also reduced in dcl4-2, although not as much as in dcl1-9 (Fig. 2A). Therefore, DCL4 may contribute in part to the formation of AtlsiRNA-1. Thus, unlike piRNAs/rasiRNA formation, that of AtlsiRNA-1 requires Dicer-like ribonuleases, which is similar to scnRNAs in Tetrahymena.
Figure 2.
Accumulation of AtlsiRNA-1 requires DCL1, DCL4, HYL1, AGO7, HST, RDR6, NRPD1a, NRPD1b, and HEN1. Level of AtlsiRNA-1 in 4-wk-old Pst (avrRpt2)-infected small RNA biogenesis mutants is shown in various dcl mutants and hyl1 (A), various ago mutants and hst-6 (B), and various rdr, pol IV mutants, sde3, and hen1 (C). Samples were collected at 15 hpi. Oligonucleotide probes complementary to AtlsiRNA-1, miR173, and TAS3 (D7 and D8) were used. U6 RNA was used for quantification and a loading control. (D) The 3′ end of AtlsiRNA-1 is modified. Gel-fractionated 20- to 50-nt small RNAs were examined by β-elimination assay. Synthetic RNA oligonucleotides (21 and 44 nt) were mixed in the samples as controls. Oligonucleotide probes complementary to AtlsiRNA-1, miR173, and synthetic oligos were used. U6 RNA was used as a control for measuring the RA (shown below). Ethidium bromide-stained tRNA is shown as a loading control.
DsRNA-binding protein HYL1 interacts with DCL1, which is important for its dicer function (Vazquez 2006). As expected, the level of AtlsiRNA-1 was greatly reduced in Pst (avrRpt2)-infected hyl1 as compared with the corresponding Nossen wild type (Fig. 2A). HASTY (HST), a homolog of exportin 5 in Arabidopsis, is thought to be involved in the export of specific miRNAs from the nucleus to the cytoplasm (Vazquez 2006). Reduction in level of AtlsiRNA-1 was also observed in the hst-6 mutant (Fig. 2B), which suggests that HST might assist in AtlsiRNA-1 processing and export.
Both miRNAs and siRNAs are incorporated into the RISC complex containing various AGO subfamily proteins to guide target mRNA degradation or translation inhibition, whereas piRNAs and scnRNAs are associated with members of Piwi subfamily proteins (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004; Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006). Arabidopsis contains 10 AGOs but no PIWI subfamily proteins (Vazquez 2006). To ascertain the involvement of AGO proteins in AtlsiRNA function, we examined the level of AtlsiRNA-1 in various AGO-deficient Arabidopsis mutants, except for ago1 and ago10, which have strong pleiotropic phenotypes and are difficult for pathogen infection. Interestingly, the level of AtlsiRNA-1 was markedly lower in ago7 (zip2, CS24282) than in the wild type upon Pst (avrRpt2) infection but was not affected in ago3, ago5, and ago9, and was even mildly induced in ago4 and ago2 (Fig. 2B). AGO7 was shown previously to be associated with TAS3 tasiRNA function (Adenot et al. 2006; Fahlgren et al. 2006; Garcia et al. 2006).
RDR proteins are involved in the amplification of small RNA by synthesizing dsRNA templates for DCL activity (Wassenegger and Krczal 2006). The level of AtlsiRNA-1 was lower in rdr6 than in the wild type and rdr2 mutant (Fig. 2C). SGS3 protein has been shown to cooperate with RDR6 to produce secondary siRNAs (Vazquez 2006). However, mutation in SGS3 has no effect on the accumulation of AtlsiRNA-1 (Fig. 2C). SDE3, a putative RNA helicase (Dalmay et al. 2001), is also involved in the formation of AtlsiRNA-1 (Fig. 2C). Arabidopsis Pol IV is required for the production of most endogenous siRNAs (Zhang et al. 2007). There are two forms of Pol IV, Pol IVa, and PolIVb, which have two distinct largest subunits, NRPD1a and NRPD1b, respectively. The level of AtlsiRNA-1 was clearly reduced upon Pst (avrRpt2) infection in both mutants, nrpd1a and nrpd1b (Fig. 2C), which indicates that Pol IV also plays a role in the formation of AtlsiRNA-1. Previous studies indicated that NRPD1a colocalizes with endogenous repeat loci and is required for small RNA formation, whereas the effect of NRPD1b on small RNA accumulation might be indirect due to destabilization of the common second largest subunit, NRPD2a, which will compromise Pol IVa activity (Li et al. 2006; Pontes et al. 2006).
AtlsiRNA-1 is modified at the 3′ terminus
One characteristic feature of small RNAs in plants is the methyltransferase HEN1-catalyzed 3′-end methylation, which protects small RNAs from terminal polyuridylation or exonucleolytic degradation (Yu et al. 2005). In mammals, miRNAs are not methylated, and only piRNAs carry a 2′-O-methyl group on the 3′-terminal ribose (Kirino and Mourelatos 2007; Ohara et al. 2007). Similarly, Drosophila rasiRNAs and zebrafish piRNAs are modified at the 3′ end (Gunawardane et al. 2007; Houwing et al. 2007). The level of AtlsiRNA-1 was markedly lower in hen1 than in the wild type (Fig. 2C), which suggests that AtlsiRNA-1 may be protected by methylation of its end. To confirm this result, we performed β-elimination analysis of lsiRNA-1 with synthetic 21- and 44-nt RNA oligos added to each reaction as controls. AtlsiRNA-1 exhibited a high level of resistance to periodate treatment, thereby confirming the modification of the 2′ or 3′ hydroxyl group at its 3′-terminal ribose (Fig. 2D). The same blot was probed with the oligos complementary to the 21- and 44-nt synthetic oligos, with shifts in the mobility of these RNA oligos clearly seen after β-elimination (Fig. 2D). The modification of the 3′-terminal hydroxyl group, together with the requirement of HEN1, suggests that AtlsiRNA-1 is likely methylated.
AtlsiRNA-1 regulates the expression of the antisense transcript AtRAP
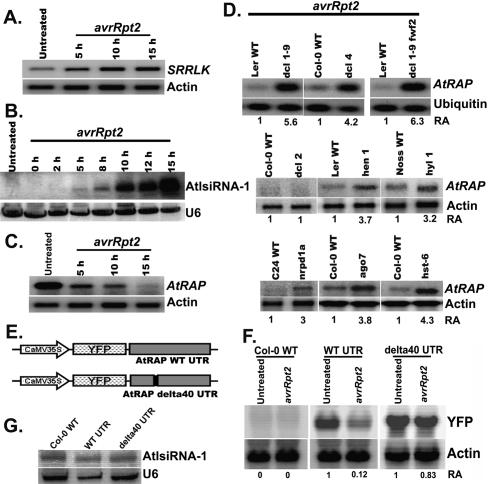
Since AtlsiRNA-1 is derived from the overlapping region of the NAT pair SRRLK/AtRAP and induced by Pst (avrRpt2), we examined the expression of these two genes in response to Pst (avrRpt2). Expression of SRRLK was clearly induced by Pst (avrRpt2) infection (Fig. 3A) but not after mock treatment (10 mM MgCl2) (Supplementary Fig. S3A). The induction pattern of AtlsiRNA-1 correlated well with that of SRRLK transcripts (Fig. 3B). AtlsiRNA-1 is derived from the exon of SRRLK and is complementary to the 3′ untranslated region (UTR) of the antisense gene AtRAP. The newly released TAIR annotation 7 shows a shorter 3′ UTR of AtRAP as compared with that in annotation 6, whereas the mRNA signatures in the MPSS database suggest that its 3′ UTR could be 950 base pairs (bp). To determine the length of AtRAP 3′ UTR, we performed RT–PCR with a forward primer (F in Fig. 4A) at the nonoverlapping region before the last intron of AtRAP and several reverse primers (R1–R6 in Fig. 4A), and confirmed that the 3′ UTR of AtRAP could extend to at least 600 bp downstream from the stop codon and 300 bp downstream from the lsiRNA-1 target site (Supplementary Fig. S3B). Quantitative RT–PCR with various RT primers reveals that >60% of the AtRAP transcripts contain the target site (Supplementary Fig. S3C). Thus, AtlsiRNA-1 likely triggers the silencing of AtRAP in cis. As expected, AtRAP expression was reduced only in response to Pst (AvrRpt2) but not mock treatment (Fig. 3C; Supplementary Fig. S3A). These observations imply that AtlsiRNA-1 might silence AtRAP upon pathogen attack.
Figure 3.
AtlsiRNA-1 down-regulates AtRAP. Expression of SRRLK (A), AtlsiRNA-1 (B), and AtRAP (C) in 4-wk-old Arabidopsis leaves at various time points post-Pst (avrRpt2) infection. DNA probes outside the overlapping region (probe A and B in Fig. 4A) were used for detecting SRRLK and AtRAP, respectively. Actin and U6 are internal controls for HMW and LMW RNA blots, respectively. (D) Level of AtRAP mRNA in selected small RNA biogenesis mutants at 15 hpi of Pst (avrRpt2). Corresponding wild-type ecotypes are included for comparison. Actin and ubiquitin were used for measuring the RA (shown below). (E) Diagram of reporter constructs carrying the YFP gene fused with the AtRAP wild-type 3′ UTR or mutated UTR with deletion of 40 nt complementary to AtlsiRNA-1. (F) Expression of YFP in transgenic Arabidopsis plants carrying reporter constructs with and without Pst (avrRpt2) infection. A YFP probe was used for Northern blot analysis. (G) Expression of AtlsiRNA-1 in the reporter transgenic lines post-Pst (avrRpt2) infection. Samples were collected at 10 hpi.
Figure 4.
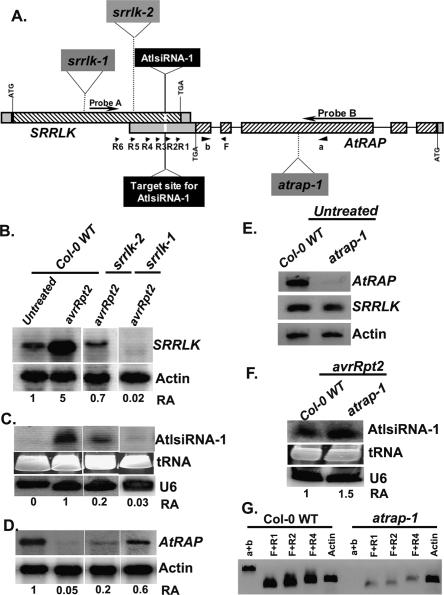
AtlsiRNA-1 formation requires the expression of sense gene SRRLK. (A) The gene structures and T-DNA insertions of SRRLK and AtRAP. (B–D) Expression of SRRLK, AtlsiRNA-1 and AtRAP in T-DNA insertion lines srrlk-1 (CS_2992) and srrlk-2 (Salk_009453). (E) Expression of SRRLK and AtRAP in untreated atrap-1 (CS_844807). (F) Level of AtlsiRNA-1 in atrap-1 at 15 hpi of Pst (avrRpt2). Probes A and B located outside the overlapping region were used for Northern blot analysis to detect SRRLK and AtRAP, respectively. U6 and actin are controls. (G) Low level of AtRAP is detected at the overlapping region in atrap-1. mRNA (1.5 μg) was used for RT with oligo dT. The expression of the 5′ region of AtRAP was examined by PCR with primers a and b. The expression of the 3′-end overlapping region of AtRAP was examined by PCR with primer F before the last intron and a series of reverse primers downstream from the last intron, R1, R2, and R4.
To confirm the effect of AtlsiRNA-1 on AtRAP mRNA degradation, we examined the transcript levels of AtRAP in RNA interference (RNAi) biogenesis mutants showing greatly reduced level of AtlsiRNA-1 in response to Pst (avrRpt2) infection. As expected, the level of AtRAP transcript was clearly elevated in dcl1-9 and dcl1-9 fwf-2 mutants (Fig. 3D). Modest induction of AtRAP was also found in dcl1-7 even without bacterial challenge in the ASRP database (http://asrp.cgrb.oregonstate.edu). Similarly, dcl4, hen1, hyl1, nrpd1a, ago7, and hst-6 mutants retained higher levels of AtRAP transcript than their respective wild types (Fig. 3D). However, like in the wild type, AtRAP was degraded in dcl2, which correlates well with the induction of AtlsiRNA-1 in dcl2 (Fig. 3D). The negative correlation between AtlsiRNA-1 and AtRAP suggests that AtlsiRNA-1 likely silences AtRAP upon avrRpt2 infection.
To further confirm that the down-regulation of AtRAP is indeed caused by the induction of AtlsiRNA-1, we designed two reporter fusion constructs carrying yellow fluorescent protein (YFP) gene fused with the wild-type or mutated 3′ UTR of AtRAP driven by a constitutive 35S promoter (Fig. 3E). The mutated 3′ UTR construct had a 40-nt deletion, which covered the region complementary to AtlsiRNA-1. Both constructs were transformed into Arabidopsis plants, and stable high-expression lines were selected for expression analysis. The level of YFP transcript was examined in transgenic plants infected with Pst (avrRpt2) at 10 hpi. Since avrRpt2 infection induces AtlsiRNA-1, which targets the AtRAP 3′ UTR, we would expect down-regulation of YFP. Northern blot analysis showed lower YFP levels in the infected YFP-WT UTR plants than in the uninfected plants (Fig. 3F). Nevertheless, 35S∷YFP-δ 40 UTR plants showed no significant change in expression of the reporter gene (Fig. 3F). The difference observed was not due to the reduced level of AtlsiRNA-1 in 35∷YFP-δ 40 UTR plants (Fig. 3G). This result suggests that AtlsiRNA-1 is responsible for the down-regulation of AtRAP.
Generation of AtlsiRNA-1 requires the sense transcript SRRLK
To determine whether generation of AtlsiRNA-1 depends on both sense and antisense transcripts of the NAT pair, we identified homozygous T-DNA insertion mutants of SRRLK (srrlk-1 for CS_2992 and srrlk-2 for Salk_009453) and AtRAP (atrap-1 for CS_844807). SRRLK is an intronless gene, and both T-DNA lines contain insertions in the coding region (Fig. 4A). The T-DNA insertion in srrlk-1 knocked out SRRLK expression, and no AtlsiRNA-1 was detected (Fig. 4B,C). srrlk-2 displayed weak induction of SRRLK and low level of AtlsiRNA-1 upon Pst (avrRpt2) infection (Fig. 4B,C). Expression of SRRLK at the overlapping region was also knocked out or knocked down in srrlk-1 and srrlk-2, respectively (Supplementary Fig. S4). Thus, generation of AtlsiRNA-1 depends on the expression of SRRLK.
In atrap-1, the T-DNA was inserted in the third exon and knocked out the expression of full-length AtRAP (Fig. 4A,E) but had no effect on the expression of AtlsiRNA-1 and SRRLK mRNA (Fig. 4E,F). However, the 3′ end of AtRAP was still expressed in the knockout line, although at a lower level than that in the control (Fig. 4G). Thus, we cannot rule out that the reduced expression of the 3′-end partial transcript may be still enough for generating lsiRNA-1.
AtlsiRNA-1 contributes to plant bacterial resistance by silencing AtRAP—a negative regulator of plant defense
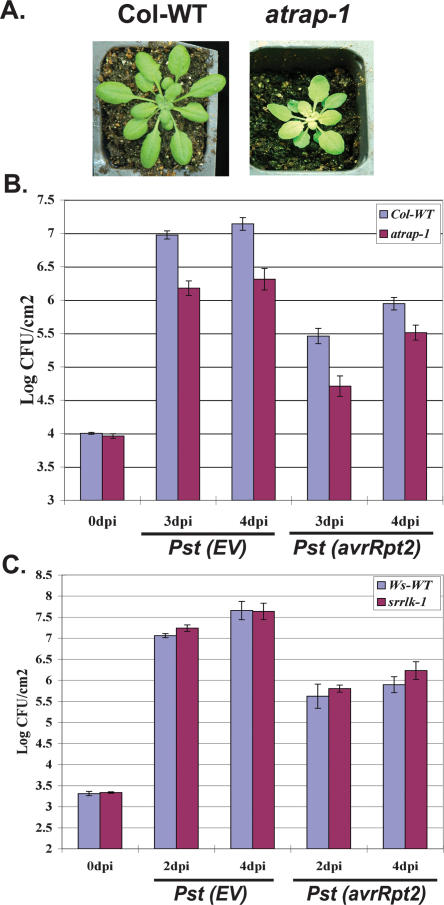
To assess the biological function of AtlsiRNA-1, we examined the knockout mutants atrap-1 and srrlk-1 for alteration in plant disease resistance. atrap-1 plants exhibited retarded growth and a photobleached phenotype (Fig. 5A). The developmental phenotype is more obvious at younger stage (within the first 4 wk post-germination). Modest but significantly less growth of both the virulent Pst (EV) and avirulent Pst (avrRpt2) was observed in atrap-1 leaves than in the wild-type control at 3–4 d post-inoculation (dpi) (Fig. 5B). Consistent results were obtained from three independent experiments. The increased resistance of atrap-1 to both virulent and avirulent strains suggests that AtRAP may be involved in basal defense and negatively regulates resistance responses. Although SRRLK is induced by Pst (avrRpt2), srrlk-1 plants displayed no significant difference in Pst (EV) and Pst (avrRpt2) growth from the wild-type Ws control (Fig. 5C). It might be because SRRLK itself plays a minor role in plant defense but rather contributes to defense indirectly by inducing AtlsiRNA-1. Alternatively, it may function redundantly with other RLKs, which is a large gene family in Arabidopsis. From these observations, we conclude that AtlsiRNA-1 may be responsible for imparting resistance against Pst (avrRpt2) infection by down-regulating a negative regulator of plant basal defense responses. We are currently investigating the molecular mechanism of AtRAP function.
Figure 5.
atrap-1 shows enhanced resistance to both virulent and avirulent bacteria. (A) atrap-1 has retarded growth and photobleached phenotype. Picture was taken at 20 d after germination. (B) Mutant atrap-1 exhibits enhanced resistance to both virulent Pst (EV) and avirulent Pst (avrRpt2). Bacterial growth was measured at 0, 3, and 4 dpi of Pst (EV) or Pst (avrRpt2) at a concentration of 2 × 105 cfu/mL. Five-week-old plants were used for the analysis. Error bars represent standard deviation of four to six replicates. Similar results were obtained in three independent experiments. (C) Mutant srrlk-1 shows no significant difference in bacteria growth as compared with Ws wild-type. Four-week-old plants were used and bacterial growth was measured at 0, 2, and 4 dpi.
AtlsiRNA-1 may promote AtRAP mRNA decapping and decay
Small RNAs induce transcriptional gene silencing by guiding DNA methylation or chromatin modification or induce post-transcriptional gene silencing by guiding target mRNA degradation and/or translation inhibition. We examined a mutant in DDM1, a gene contributing to maintenance of RNA-directed DNA methylation and responsible for the majority of the genomic cytosine methylation (Jeddeloh et al. 1999). The level of AtlsiRNA-1 and AtRAP mRNA did not change in ddm1-2 after Pst (avrRpt2) infection as compared with the wild-type control (Fig. 6A), which implies that silencing of AtRAP might occur mainly at the post-transcriptional level. The reduction of AtRAP mRNA level in response to Pst (avrRpt2) (Fig. 3C,D) indicates that the major mode of action of AtlsiRNA-1 is to trigger mRNA degradation rather than translation inhibition. In plants, miRNAs and siRNAs mainly direct endonucleolytic cleavage of target mRNAs by AGO slicer activity on perfect or nearly perfect base pairing. However, we could not detect any cleavage product of AtRAP by Northern blot analysis or 5′ RLM-RACE (data not shown). These results point to the likelihood of other mechanisms involved in the mode of action of AtlsiRNA-1, although cleavage of AtRAP could not be completely ruled out because of the low level of expression of AtRAP and the limited sensitivity of our assays.
Figure 6.
The level of AtRAP mRNA is not affected in the ddm1 mutant but is accumulated more in decapping complex mutants tdt-2mc and vcs-1 and the 5′-to-3′ exoribonuclease mutant xrn4-5 than in wild-type controls. (A) Expression of lsiRNA-1 and AtRAP in ddm1 at 15 hpi of Pst (avrRpt2). (B) Expression of AtRAP and lsiRNA-1 in decapping complex mutants vcs-1 and tdt-2mc at 15 hpi of Pst (avrRpt2). AtRAP and AtHB15 were detected by semiquantitative RT–PCR because not enough mRNA could be obtained. PCR products after 25 cycles (for AtRAP and AtHB15) and 20 cycles (for ubiquitin) were analyzed by Southern blot analysis. (Bottom panel) AtlsiRNA-1 was examined by Northern blot analysis. (C,D) Degradation of AtRAP in xrn4-5 mutant (C) and XRN2 T-DNA knockout mutant (D) examined at 6 hpi of Pst (avrRpt2) after cordycepin treatment. At1g78080 transcript is a known target of XRN4. Specific probes for detecting AtRAP, At1g78080, and actin were used.
Several lines of evidence in animal systems suggest that small RNAs can also accelerate target mRNA degradation by cleavage-independent mechanisms such as deadenylation and/or decapping followed by 5′-to-3′ degradation (Behm-Ansmant et al. 2006; Giraldez et al. 2006; Valencia-Sanchez et al. 2006; Wu et al. 2006). Usually, on Northern blot analysis, deadenylation of mRNA shows increased mobility over time because of the shortened poly(A) tail (Behm-Ansmant et al. 2006; Wu et al. 2006). We did not detect any change in the size of AtRAP mRNA during degradation over the time course of 15 hpi of Pst (avrRpt2) infection (Fig. 3C). Whether deadenylation is involved in AtRAP mRNA decay cannot be easily tested because the available Arabidopsis deadenylase mutant is embryo lethal (Chiba et al. 2004).
Removal of the 5′ cap from the mRNA is an important trigger for mRNA decay. Decapping enzymes are localized in processing (P)-bodies, where mRNA degradation occurs (Liu et al. 2005). Some AGO proteins are associated with P-bodies and interact with P-body proteins, including decapping enzymes, for mRNA degradation in human, worm, and fly (Ding et al. 2005; Liu et al. 2005; Sen and Blau 2005; Behm-Ansmant et al. 2006). In Arabidopsis, the decapping complex comprising DCP1, DCP2, and VARICOSE (VCS) was shown to localize in putative P-bodies. Mutations in DCP1, DCP2, and VCS affect the turnover of a group of mRNAs but not all mRNAs, and are seedling lethal (Xu et al. 2006; Goeres et al. 2007). To examine the effect of mRNA decapping on AtRAP degradation, we took the advantage of the temperature-dependent mutants vcs-1 (in the Ler accession) and DCP2 mutant tdt MC2 (Col tdt mutant after two sequential crosses to Ler) (Goeres et al. 2007), which can produce broad pointed leaves when grown at 16°C and allow for conducting bacterial infection assay. Although AtHB15 level was not changed in vcs-1 and tdt MC2 after infection (Fig. 6B), AtRAP mRNA showed a more than sixfold increase in vcs-1 and tdt MC2 than in the Pst (avrRpt2)-infected wild type (Fig. 6B), which almost completely inhibited the degradation of AtRAP mRNA after infection as compared with the untreated wild type (Fig. 6B). These results suggest that the degradation of AtRAP mRNA upon Pst (avrRpt2) infection largely depends on the decapping complex. The inhibition of AtRAP degradation was not due to the alteration of the AtlsiRNA-1 level. In fact, the AtlsiRNA-1 level was even higher in tdt MC2 than in the Ler control (Fig. 6B). These data, together with the result that lsiRNA-1 target site deletion could largely restore the expression of the reporter YFP, suggest that AtlsiRNA-1 might induce silencing of AtRAP through RNA decapping.
Decapped mRNAs are subjected to degradation by 5′-to-3′ exoribonucleases. Arabidopsis contains three 5′-to-3′ exoribonucleases, XRN2, XRN3, and XRN4, which are homologous to yeast XRN1. XRN4 recognizes the 5′ end of decapped or cleaved transcripts for degradation. Mutation in XRN4 affects the mRNA levels of only a few genes, causes no obvious phenotype, and promotes RDR-mediated gene silencing (Gazzani et al. 2004; Souret et al. 2004). We detected AtRAP transcript accumulated to a higher level in the xrn4-5 mutant than in the wild type and XRN2 knockout line (Salk_114258) that we isolated (Fig. 6C,D), which suggests that AtRAP degradation requires XRN4.
Discussion
Here, we identified a novel class of 30- to 40-nt-long noncoding RNAs in Arabidopsis, AtlsiRNAs. Unlike animal piRNAs, which are mainly derived from transposons, retroelements, and repetitive loci, and are only present in germline cells, the AtlsiRNAs we identified are generated from gene-coding regions, present in various tissues, and are induced by pathogen attack or under specific growth conditions. Further study of one of the Pst (avrRpt2) infection-induced lsiRNAs, AtlsiRNA-1, revealed that its generation depends on DCL1 and DCL4 but not DCL2 and DCL3. We predict that AtlsiRNA-1 is primarily generated by DCL1 in response to Pst (avrRpt2) and then undergoes secondary amplification by Pol IV, RDR6, SDE3, and DCL4, which explains why these mutants had less effect on AtlsiRNA-1 accumulation than dcl1 mutants did.
Although currently known small RNAs generated by DCL1 in Arabidopsis have a rather strict size range of 20- to 22-nt, several studies using deep parallel sequencing have identified heterogeneous miRNAs with various lengths of 20- to 24-nt within many individual miRNA loci (Rajagopalan et al. 2006; Fahlgren et al. 2007). As well, Tetrahymena DCL1 can produce scnRNAs of 26- to 31-nt (Mochizuki and Gorovsky 2005). Therefore, Arabidopsis DCL1 and DCL4 could possibly generate lsiRNAs. HYL1 has been shown to play an important role in the “dicer” function of DCL1. HYL1 mutation can also greatly reduce the level of AtlsiRNA-1. Interaction of DCL1 with other proteins, including HYL1 and other unidentified components, may influence the size of the small RNAs. Since AtlsiRNA-1 is induced only by Pst (avrRpt2) challenge, it is possible that bacterial proteins, such as avrRpt2, might directly or indirectly interfere with DCL1 and alter its function in generating a specific class of small RNAs. Alternatively, DCL1 and DCL4 may be responsible for cleavage of only one end of the lsiRNA-1, and other proteins with nuclease activity may process the other end. The biogenesis of AtlsiRNA-1 may involve the slicer activity of AGO7, just like the roles of AGO3 and Aubergine PIWI subfamily proteins in the biogenesis of piRNAs/rasiRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). However, the enzymatic slicer activity of AGO7 has yet to be demonstrated. It is also possible that generation of lsiRNA-1 may require the action of other undetermined nucleases, like the roles of Zucchini and Squash proposed in the formation of rasiRNAs (Pane et al. 2007). However, the close homologs of Zucchini and Squash have not been identified in Arabidopsis.
Our results suggest that AtlsiRNA-1 might trigger AtRAP silencing by destabilizing its mRNA through decapping and XRN4-mediated 5′-to-3′ degradation. Most plant miRNAs and endogenous siRNAs studied so far mainly cause cleavage of target mRNA or translational inhibition; small RNA-induced mRNA instability has not yet been reported in plants. However, inducing cleavage-independent degradation of target mRNA is a rather common mode of action of small RNAs in animal systems (Valencia-Sanchez et al. 2006). Several animal AGO proteins are associated with decapping enzymes and are localized in P-bodies (Ding et al. 2005; Liu et al. 2005; Sen and Blau 2005; Behm-Ansmant et al. 2006). AtlsiRNA-1 requires AGO7 for its biogenesis and function. AGO7 was shown to be involved in the function of TAS3 tasiRNAs, but its functional mechanism and subcellular localization has not been shown. We speculate that after Pst (avrRpt2) infection, lsiRNA-1 will associate with an AGO7-containing RISC complex and bring its target mRNA to a P-body for decapping and degradation. Future studies to demonstrate the subcellular localization of AGO7 in response to Pst (avrRpt2) infection and the association of AtlsiRNAs with AGO7 will add strong evidence to this hypothesis.
Functional analysis of AtRAP revealed its negative role in plant disease resistance. Plants have evolved a sophisticated mechanism to control its resistance signaling network (Chisholm et al. 2006; Jones and Dangl 2006). To be cost effective, the resistance pathways need to be “off” under normal growth conditions and should be quickly turned “on” under pathogen attack. AtRAP may function in controlling the resistance responses under the normal condition. However, the suppression of resistance responses by AtRAP can be quickly lifted by the induction of AtlsiRNA-1 in response to incompatible Pst (avrRpt2) infection, which leads to rapid degradation of AtRAP mRNA. AtRAP may play a role in basal defense because it affects both compatible and incompatible interaction between Pst and plants. We are in the process of exploring the functional mechanism of AtRAP in plant defense.
The discovery of AtlsiRNAs has added to the complexity of the small RNA world in plants. Some of these small RNAs are induced in response to pathogen attack, and are therefore implicated in disease resistance in plants. AtlsiRNAs may play diverse functions in plants, including regulation of pathogen-responsive genes. Identification and characterization of more lsiRNAs will shed light on the biogenesis mechanisms and regulatory functions of these small RNAs in plants.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants were grown in a controlled growth room at 23°C ± 1°C with a 12-h light/12-h dark photoperiod. tdt-MC2 and vcs-1 plants along with Ler were grown for 6–8 wk at 16°C and acclimated for 2 d at 24°C before bacteria infiltration. All experiments were performed on 4-wk-old Arabidopsis plants. We thank the following researchers for the seeds used in this study: dcl1-9 and hen1-1 (Xumei Chen, University of California at Riverside, Riverside, CA); hyl1 (Nina Fedoroff, The Huck Institute of Life Science, Pennsylvania State University, University Park, PA); rdr2-1, dcl2-1, dcl3-1, and dcl4-2 (Jim Carrington, Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR); ago4 (Steve Jacobsen, University of California at Los Angeles, Los Angeles, CA); sde1/rdr6, sde3, and sde4/nrpd1a (David Baulcombe, Sainsbury Laboratory, Norwich, UK); ago2 (Zhixin Xie, Texas Tech University, Lubbock, TX); ago3 (John Clarke, John Innes Center, Norwich, UK); nrpd1b (Thierry Lagrange, Université de Perpignan, Cedex, France); sgs3 (Herve Vaucheret, Institut National de la Recherche Agronomique, Versailles, France); ddm1-2 (Eric Richards, Wash-ington University, St. Louis, MO); tdt-MC2 and vcs-1 (Leslie E. Sieburth, University of Utah, Salt Lake City, UT); xrn4 (Pam Green, University of Delaware, Newark, DE). Ago 7 (zip 2, CS24282), hasty-6 (CS24279), ago5 (Salk_063806), ago9 (Salk_127358), SRRLK (Salk_09453 and CS2992), AtRAP (CS844807), and xrn2 (Salk_114258) were ordered from ABRC, Ohio. The homozygous plants were identified by PCR, and the expression of the genes was examined by RT–PCR or Northern blot analysis. ago5 (Salk_063806), ago9 (Salk_127358), and xrn2 (Salk_114258) are knockout mutants. All these mutants were in the Columbia (Col-0), Landsberg erecta (Ler), Nossen-0 (Noss), Wassilewskija (Ws) or C24 genetic backgrounds as indicated in the text and figures, respectively.
Plasmid DNA constructs and generation of transgenic Arabidopsis plants
To obtain the 35S∷YFP-AtRAPUTR constructs, a 950-bp fragment right after the stop codon of AtRAP was amplified with the AtRAP-UTR primers (Supplementary Table S1) and cloned into the plant expression Gateway destination vector pEG104 (Earley et al. 2006) containing an N-terminal YFP protein tag driven by the constitutive 35S cauliflower mosaic virus promoter. To clone the 3′ UTR insensitive to small RNAs, 40 bases complementary to the AtlsiRNA-1 (24-nt MPSS sequence [uppercase and labeled pink in Supplementary Fig. S1] and 16 nt on the 5′ end) in the 3′ UTR region of AtRAP were deleted, and the fragment was cloned in pEG104 (AtRAP-δ40 UTR) as described above. Agrobacterium tumefaciens strain GV3101 was used for Col-0 wild-type transformation.
RNA Northern blot analysis
RNA extraction and small RNA Northern analysis were carried out as described (Katiyar-Agarwal and Jin 2007). Imagequant version 2.1 software was used for RA quantification (GE Healthcare Life Sciences).
For high-molecular-weight (HMW) RNA blot analysis, we mainly used total RNA for SRRLK detection and enriched mRNA isolated by the use of Poly(A) tract isolation system (Promega) for AtRAP detection because AtRAP expression is very low. About 3–5 μg of mRNA were resolved on 1.2% agarose formaldehyde-MOPS gel. The RNA was transferred to Duralon-UV membrane (Stratagene) by capillary blotting. cDNA probes were labeled with α-32P dCTP by the Prime-it II random primer labeling system (Stratagene). RT–PCR was used to amplify specific probes (Fig. 4A; Supplementary Table S1).
RT–PCR
For checking the expression levels of AtRAP, first-strand cDNA was synthesized from 1–2 μg of mRNA with oligo dT because AtRAP expression level is very low. Then PCR was performed with various forward and reverse primers (Fig. 4A; Supplementary Table S1). The PCR fragments were confirmed by sequencing. Real-time RT–PCR was used to check the expression level of various AtRAP transcripts. AtRAP-specific primers (R2, R4, R5, and R6) were used for RT in addition to the oligo dT primer. All the cDNA was subjected for PCR using primers F and b to ensure the same PCR efficiency. SRRLK-specific primer SRRLK-stop R was used for RT to check the expression of the overlapping region of SRRLK.
Small RNA cloning
Cloning of small RNA was performed as described previously (Sunkar and Zhu 2004). Briefly, small RNAs isolated from Pst (avrRpt2)-infected Arabidopsis leaves were size-fractionated, purified, and ligated sequentially to 5′- and 3′-end RNA oligonucleotide adapters. RT involved a primer specific for the 3′ adaptor. Since a 24-nt sequence was already known for AtlsiRNA-1, the flanking sequences were cloned by PCR with a combination of primers specific for the 5′-end adaptor and 24-nt MPSS reverse sequence, as well as the 3′-end adaptor and 24-nt MPSS forward sequence. The resulting PCR products were cloned in pGemT-Easy vector (Promega), and ligation products were transformed in bacteria. Plasmids isolated from different bacterial clones were sequenced.
β-Elimination reaction of small RNAs
Small RNAs from Pst (avrRpt2)-infected leaves were size-fractionated from denaturing polyacrylamide gel. The purified fraction of 20- to 50-nt small RNAs was incubated in a solution containing 10 mM HEPES (pH 7.0) and 250 mM sodium periodate for 30 min at 22°C. An equal volume of formamide loading dye was added to the samples, followed by incubation for 45 min at 99°C. The reaction mixture was resolved on a 19-cm-long, 13% denaturing polyacrylamide gel. An equal amount of untreated RNA was also loaded onto the gel for comparison. To check the completion of sodium periodate treatment, RNA oligos of known sequence (21- and 44-nt synthetic RNA oligo) were also mixed with the small RNA during β-elimination reaction, and the blots were probed with end-labeled oligos complementary to the synthetic oligos and AtlsiRNA-1.
Bacteria infection
For RNA analysis, 4-wk-old Arabidopsis leaves were infiltrated with Pst DC3000 carrying EV (pVSP61) or avr genes avrRpt2, avrRps4, avrPphB, or avrRpm1 at a concentration of 2 × 107 colony-forming units (cfu) per milliliter. Infected leaves were harvested at various time points before hypersensitive responses occur. For YFP-UTR experiments, samples were collected at 10 hpi. For xrn2 and xrn4 mutant experiments, leaves from 4-wk-old Arabidopsis plants were excised and floated on half-strength MS medium containing 0.005% Silwett-L77. Cordycepin (Sigma Chemicals) was added to the medium at a final concentration of 300 μg/mL for pretreatment for 2.5 h. Leaves were then infiltrated with 2 × 107 cfu/mL Pst (avrRpt2), and the tissue was harvested 6 hpi. As a control, leaves were floated on MS medium containing Silwett-L77 for 8.5 h.
For Flg22 treatment, 1-wk-old cell suspension culture of Arabidopsis was treated with 50 nM Flg22 peptide and harvested at different time points.
For bacterial growth assay in atrap-1, Pst (EV) or Pst (avrRpt2) were infiltrated into 5-wk-old Arabidopsis leaves at a concentration of ∼2 × 105 cfu/mL. Leaf samples were collected by a cock borer, and the bacterial titer were determined by plating and counting the number of colonies at 0, 3, and 4 dpi. Four to six replicates were taken for each set of experiments. The experiment was repeated three times and similar results were obtained. For srrlk-1, the same pathogen assay was performed on 4-wk-old plants at 0, 2, and 4 dpi.
Acknowledgments
We thank Shou-Wei Ding, Xuemei Chen, and Jian-Kang Zhu for stimulating discussion and providing seeds of various mutants, and James Borneman for access to a real-time Icycler in his laboratory. We also thank David Baulcombe, Jim Carrington, Leslie E. Sieburth, Pam Green, Nina Fedoroff, Herve Vaucheret, Steve Jacobsen, John Clarke, Thierry Lagrange, and Eric Richards for providing seeds of various genotypes. This work was supported by an NSF Career Award MCB-0642843, a University of California Discovery Grant Bio06-10566, and an AES-CE Research Allocation Award PPA-7517H.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1595107
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H., Boutet S., Bouche N., Gasciolli V., Vaucheret H., Bouche N., Gasciolli V., Vaucheret H., Gasciolli V., Vaucheret H., Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Naumova N.M., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A., Naumova N.M., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A., Tulin A.V., Vagin V.V., Rozovsky Y.M., Gvozdev V.A., Vagin V.V., Rozovsky Y.M., Gvozdev V.A., Rozovsky Y.M., Gvozdev V.A., Gvozdev V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Snyder B., Gaasterland T., Meyer J., Tuschl T., Gaasterland T., Meyer J., Tuschl T., Meyer J., Tuschl T., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Kuramochi-Miyagawa S., Nakano T., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Doerks T., Stark A., Bork P., Izaurralde E., Stark A., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. MRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J.H., Verslues P.E., Sunkar R., Zhu J.K., Zhu J.H., Verslues P.E., Sunkar R., Zhu J.K., Verslues P.E., Sunkar R., Zhu J.K., Sunkar R., Zhu J.K., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Dus M., Kellis M., Sachidanandam R., Hannon G.J., Kellis M., Sachidanandam R., Hannon G.J., Sachidanandam R., Hannon G.J., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Johnson M.A., Lidder P., Vogel J.T., van Erp H., Green P.J., Johnson M.A., Lidder P., Vogel J.T., van Erp H., Green P.J., Lidder P., Vogel J.T., van Erp H., Green P.J., Vogel J.T., van Erp H., Green P.J., van Erp H., Green P.J., Green P.J. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J., Coaker G., Day B., Staskawicz B.J., Day B., Staskawicz B.J., Staskawicz B.J. Host–microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Horsefield R., Braunstein T.H., Baulcombe D.C., Horsefield R., Braunstein T.H., Baulcombe D.C., Braunstein T.H., Baulcombe D.C., Baulcombe D.C. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., Spencer A., Morita K., Han M., Morita K., Han M., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S., Opper K., Juehne T., Song K.M., Pikaard C.S., Juehne T., Song K.M., Pikaard C.S., Song K.M., Pikaard C.S., Pikaard C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C., Dvorak S.K., Alexander A.L., Carrington J.C., Alexander A.L., Carrington J.C., Carrington J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Law T.F., Grant S.R., Dangl J.L., Grant S.R., Dangl J.L., Dangl J.L., et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A., Collier S.A., Byrne M.E., Martienssen R.A., Byrne M.E., Martienssen R.A., Martienssen R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R., Lawrenson T., Woodward C., Headon D., Sablowski R., Woodward C., Headon D., Sablowski R., Headon D., Sablowski R., Sablowski R. A link between rnRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G., Carmell M., Sachidanandam R., Hannon G., Carmell M., Hannon G., Carmell M., Carmell M. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goeres D.C., Van Norman J.M., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E., Van Norman J.M., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E., Fauver N.A., Spencer M.L., Sieburth L.E., Spencer M.L., Sieburth L.E., Sieburth L.E. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H.F., Beyret E., Wang Z., Lin H.F., Wang Z., Lin H.F., Lin H.F. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Givan S., Smith D., Carrington J.C., Kasschau K.D., Smith D., Carrington J.C., Kasschau K.D., Carrington J.C., Kasschau K.D., Kasschau K.D. ASRP: The Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz S.R., Cumbie J.S., Kasschau K.D., Lohmann J.U., Carrington J.C., Weigel D., Schmid M., Cumbie J.S., Kasschau K.D., Lohmann J.U., Carrington J.C., Weigel D., Schmid M., Kasschau K.D., Lohmann J.U., Carrington J.C., Weigel D., Schmid M., Lohmann J.U., Carrington J.C., Weigel D., Schmid M., Carrington J.C., Weigel D., Schmid M., Weigel D., Schmid M., Schmid M. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 2007;144:1247–1255. doi: 10.1104/pp.107.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., Filippov D.V., Blaser H., Raz E., Moens C.B., Blaser H., Raz E., Moens C.B., Raz E., Moens C.B., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J., Stokes T.L., Richards E.J., Richards E.J. Maintenance of genomic methylation requires a SW12/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Cumbie J.S., Givan S.A., Carrington J.C., Givan S.A., Carrington J.C., Carrington J.C. Genome-wide profiling and analysis of ArabidopsissiRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Jin H., Jin H. Discovery of pathogen-regulated small RNAs in plants. Methods Enzymol. 2007;427:215–227. doi: 10.1016/S0076-6879(07)27012-0. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A., Zhu J.K., Staskawicz B.J., Jin H.L., Morgan R., Dahlbeck D., Borsani O., Villegas A., Zhu J.K., Staskawicz B.J., Jin H.L., Dahlbeck D., Borsani O., Villegas A., Zhu J.K., Staskawicz B.J., Jin H.L., Borsani O., Villegas A., Zhu J.K., Staskawicz B.J., Jin H.L., Villegas A., Zhu J.K., Staskawicz B.J., Jin H.L., Zhu J.K., Staskawicz B.J., Jin H.L., Staskawicz B.J., Jin H.L., Jin H.L. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y., Mourelatos Z., Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Nakano T., Bartel D.P., Kingston R.E., Bartel D.P., Kingston R.E., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee S.R., Collins K., Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., Chan S.W.L., Lagrange T., Pikaard C.S., Jacobsen S.E., Lagrange T., Pikaard C.S., Jacobsen S.E., Pikaard C.S., Jacobsen S.E., Jacobsen S.E. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Liu J.D., Valencia-Sanchez M.A., Hannon G.J., Parker R., Valencia-Sanchez M.A., Hannon G.J., Parker R., Hannon G.J., Parker R., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Kasschau K.D., Rector M.A., Carrington J.C., Kasschau K.D., Rector M.A., Carrington J.C., Rector M.A., Carrington J.C., Carrington J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Haudenschild C.D., Meyers B.C., Green P.J., Meyers B.C., Green P.J., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A., Gorovsky M.A. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A., Gorovsky M.A. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A., Fine N.A., Fujisawa T., Gorovsky M.A., Fujisawa T., Gorovsky M.A., Gorovsky M.A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D., Dharmasiri N., Estelle M., Voinnet O., Jones J.D., Estelle M., Voinnet O., Jones J.D., Voinnet O., Jones J.D., Jones J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Ueda H., Miyauchi K., Suzuki T., Miyauchi K., Suzuki T., Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schüpbach T., Wehr K., Schüpbach T., Schüpbach T. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Vitins A., Jacobsen S.E., Pikaard C.S., Jacobsen S.E., Pikaard C.S., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P., Vaucheret H., Trejo J., Bartel D.P., Trejo J., Bartel D.P., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Miyoshi K., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophilagenome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Souret F.F., Kastenmayer J.P., Green P.J., Kastenmayer J.P., Green P.J., Green P.J. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Crevillen P., Liu F.Q., Ecker J.R., Jerzmanowski A., Dean C., Crevillen P., Liu F.Q., Ecker J.R., Jerzmanowski A., Dean C., Liu F.Q., Ecker J.R., Jerzmanowski A., Dean C., Ecker J.R., Jerzmanowski A., Dean C., Jerzmanowski A., Dean C., Dean C. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. 2007;104:3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C.J., Seitz H., Gvozdev V., Zamore P.D., Sigova A., Li C.J., Seitz H., Gvozdev V., Zamore P.D., Li C.J., Seitz H., Gvozdev V., Zamore P.D., Seitz H., Gvozdev V., Zamore P.D., Gvozdev V., Zamore P.D., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J.D., Hannon G.J., Parker R., Liu J.D., Hannon G.J., Parker R., Hannon G.J., Parker R., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vazquez F. Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wang X.J., Gaasterland T., Chua N.H., Gaasterland T., Chua N.H., Chua N.H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Krczal G., Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wu L.G., Fan J.H., Belasco J.G., Fan J.H., Belasco J.G., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J.Y., Niu Q.W., Chua N.H., Yang J.Y., Niu Q.W., Chua N.H., Niu Q.W., Chua N.H., Chua N.H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z.Y., Li J.J., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M., Yang Z.Y., Li J.J., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M., Li J.J., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M., Minakhina S., Yang M.C., Padgett R.W., Steward R., Chen X.M., Yang M.C., Padgett R.W., Steward R., Chen X.M., Padgett R.W., Steward R., Chen X.M., Steward R., Chen X.M., Chen X.M. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Haley B., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Henderson I.R., Lu C., Green P.J., Jacobsen S.E., Henderson I.R., Lu C., Green P.J., Jacobsen S.E., Lu C., Green P.J., Jacobsen S.E., Green P.J., Jacobsen S.E., Jacobsen S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]