Abstract
In eukaryotes, shortening of the 3′-poly(A) tail is the rate-limiting step in the degradation of most mRNAs, and two major mRNA deadenylase complexes—Caf1–Ccr4 and Pan2–Pan3—play central roles in this process, referred to as deadenylation. However, the molecular mechanism triggering deadenylation remains elusive. Previously, we demonstrated that eukaryotic releasing factor eRF3 mediates deadenylation and decay of mRNA in a manner coupled to translation termination. Here, we report the mechanism of mRNA deadenylation. The eRF3-mediated deadenylation is catalyzed by both Caf1–Ccr4 and Pan2–Pan3. Interestingly, translation termination complexes eRF1–eRF3, Pan2–Pan3, and Caf1–Ccr4 competitively interact with polyadenylate-binding protein PABPC1. In each complex, eRF3, Pan3, and Tob, respectively, mediate PABPC1 binding, and a combination of a PAM2 motif and a PABC domain is commonly utilized for their contacts. A translation-dependent exchange of eRF1–eRF3 for the deadenylase occurs on PABPC1. Consequently, PABPC1 binding leads to the activation of Pan2–Pan3 and Caf1–Ccr4. From these results, we suggest a mechanism of mRNA deadenylation by Pan2–Pan3 and Caf1–Ccr4 in cooperation with eRF3 and PABPC1.
[Keywords: Translation termination, deadenylation, eRF3, PABPC1]
Control of mRNA decay is a fundamentally important step in determining the amount of protein produced from the mRNA by translation, and mRNA decay is intimately linked to and regulated by translation. In eukaryotes, decay of most mRNAs is initiated by shortening of the poly(A)-tail at the 3′ end (Shyu et al. 1991; Muhlrad and Parker 1992), which is followed by two general pathways: the 5′-to-3′ and 3′-to-5′ exonucleolytic degradation pathways. The poly(A) tail-shortening process, referred to as deadenylation, is the first and rate-limiting step, and is also the most efficient step in controlling mRNA decay (Decker and Parker 1993).
Two distinct enzyme complexes involved in the deadenylation process have been identified in yeast (Daugeron et al. 2001; Tucker et al. 2001) and mammals (Yamashita et al. 2005). One major cytoplasmic deadenylase complex is Ccr4–Caf1, which was originally implicated in the control of transcription (Denis and Chen 2003). As suggested by the significant homology with known 3′-to-5′ exonucleases, both Ccr4 and Caf1 have catalytic activity of the deadenylase (Chen et al. 2002; Tucker et al. 2002; Thore et al. 2003; Viswanathan et al. 2004; Bianchin et al. 2005). Another major deadenylase complex, PAN, was identified in yeast (Sachs and Deardorff 1992) and in mammal (Uchida et al. 2004). PAN is a heterodimeric protein consisting of catalytic Pan2 and regulatory Pan3 subunits (Brown et al. 1996; Uchida et al. 2004). In contrast to Ccr4–Caf1, PAN is characterized by the requirement of PABPC1 for its deadenylating activity: Pan2 is activated by binding to PABPC1 through the interaction with Pan3 (Mangus et al. 2004; Uchida et al. 2004).
Since deadenylation is the rate-limiting step for mRNA decay, the enzymatic activities of the two mRNA deadenylases constitute a major target for the control of mRNA decay. Recent findings demonstrated that selective recruitment of deadenylating enzymes to the substrate mRNA is involved in the decay mechanism for specific mRNAs (Semotok et al. 2005; Finoux and Seraphin 2006; Goldstrohm et al. 2006). However, the general mechanism triggering deadenylation of mRNA has not been elucidated. Several lines of evidence have revealed a direct link between translation termination and mRNA decay.
In eukaryotes, translation termination is governed by heterodimeric releasing factors eRF1 and eRF3 (Frolova et al. 1994; Stansfield et al. 1995; Zhouravleva et al. 1995). eRF1 directly recognizes all three stop codons in the ribosomal A site to release the completed polypeptide chain from the ribosome (Frolova et al. 1994), and eRF3 stimulates the termination reaction in a GTP-dependent manner (Zhouravleva et al. 1995). The eRF3 gene is conserved from yeast to mammals. In the yeast Saccharomyces cerevisiae, eRF3 is encoded by the essential gene SUP35 (Stansfield et al. 1995), and the mammalian eRF3 gene GSPT was first identified by its ability to complement the temperature-sensitive growth arrest phenotype of a sup35/gst1-1 mutant S. cerevisiae that is defective in the G1-to-S-phase transition (Hoshino et al. 1989). Later, two subtypes of GSPT genes, GSPT1 and GSPT2, were identified (Hoshino et al. 1998). eRF3 consists of two domains: the unique N-terminal region (N domain) and the C-terminal region (C domain), which contains eEF1A-like GTP-binding motifs. The C domain of eRF3 is sufficient to fulfill the functions of eRF3 in termination (Zhouravleva et al. 1995).
In addition to translation termination, eRF3 functions in translation termination-coupled events. We showed previously that eRF3 interacts with poly(A)-binding protein (PABPC1) through its N domain (Hoshino et al. 1999). The interaction is evolutionarily conserved from yeast to mammals (Hoshino et al. 1999; Kozlov et al. 2001; Cosson et al. 2002; Uchida et al. 2002; Hosoda et al. 2003). In yeast, eRF3 regulates the initiation of normal mRNA decay at the poly(A) tail-shortening step through the interaction with PABPC1 in a manner coupled to translation termination (Hosoda et al. 2003). Recently, Tpa1—which interacts with eRF1/eRF3 and affects translation termination, deadenylation, and mRNA decay—was identified (Keeling et al. 2006). On the other hand, in nonsense codon-containing mRNA, translation termination is thought to occur via the termination complex eRF1/eRF3 at the premature termination codon. eRF1/eRF3 associates with Upf1/Upf2/Upf3 to form a “surveillance complex” and triggers rapid mRNA decay via the nonsense-mediated mRNA decay (NMD) pathway (Wang et al. 2001; Kobayashi et al. 2004; Kashima et al. 2006).
In this study, we examined the mechanism of mRNA deadenylation and obtained evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. We demonstrate that eRF3-mediated deadenylation is catalyzed by both of the two major mRNA deadenylase complexes in yeast and humans. Interestingly, translation termination complex eRF1–eRF3 and the two mRNA deadenylase complexes competitively bind to the PABC domain of PABPC1. The PAM2 motifs found in both eRF3 and the two deadenylase complexes are responsible for their binding to PABPC1 and mRNA deadenylation. The termination complex and the deadenylase complex are exchanged on PABPC1 in a translation-dependent manner. Recruitment of the two deadenylase complexes to PABPC1 leads to the activation of both enzymes. These results provide a molecular basis for the mechanism of mRNA deadenylation.
Results
The two major mRNA deadenylases, Pan2–Pan3 and Caf1–Ccr4, are involved in the eRF3-mediated deadenylation of mRNA in S. cerevisiae
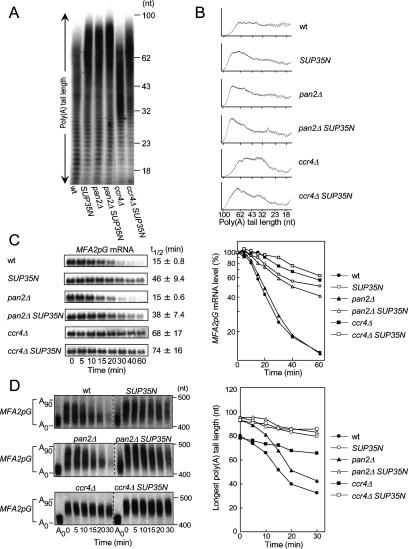
Using S. cerevisiae as a model organism, we showed previously that eRF3 mediates deadenylation and decay of normal mRNA in a manner coupled to translation termination (Hosoda et al. 2003). To identify the deadenylases involved in the eRF3-mediated deadenylation, we first examined the possible involvement of the two major mRNA deadenylases, Pan2–Pan3 and Caf1–Ccr4, by comparing phenotypes in strains pan2Δ and ccr4Δ, and a strain in which the N-terminal domain of eRF3 (SUP35N) was overexpressed. In this study, we used the strain overexpressing SUP35N, since this strain and the eRF3 mutant strains sup35ΔN and sup35ts show the same phenotypes regarding mRNA deadenylation and decay (Hosoda et al. 2003). We first analyzed the poly(A) tail length of total steady-state mRNA in vivo. After labeling with [32P]pCp, total RNA was digested with RNaseA, and the remaining intact poly(A) tails were resolved on polyacrylamide gels (Fig. 1A,B). As reported previously, the overexpression of SUP35N led to an increase in bulk mRNA poly(A) tail lengths (Hosoda et al. 2003). Similarly, a deletion in PAN2 also resulted in an increase in the average poly(A) tail length. PhosphorImager analysis showed that the average length increased ∼20 nucleotides (nt) in both strains compared with the wild-type strain. The changes in poly(A) tail length on mRNA in pan2Δ overexpressing SUP35N were comparable with those found in either pan2Δ or the SUP35N-overexpressing strain. Thus, the deadenylation defect in the SUP35N-overexpressing strain was not exacerbated by pan2Δ. These results suggest that Pan2 is involved at least in part in eRF3-mediated deadenylation.
Figure 1.
Comparison of the length of poly(A) tails and decay rates in S. cerevisiae. (A) pKT10 or pKT10-SUP35N, which overexpresses the N domain of eRF3 (SUP35N) under the control of GAL1 UAS, was introduced into wild-type, pan2Δ, and ccr4Δ strains. Total RNAs at the steady-state level from the strains were 3′-labeled with [32P]pCp and digested with RNaseA. The remaining mRNA poly(A) tails were separated on a 12% polyacrylamide–7 M urea gel and visualized by autoradiography. (B) The resulting autoradiograph from A was analyzed by densitometric tracing. (C) p9L-MFA2pG was introduced into the same strains as used in A. After transcriptional repression by adding doxycycline, the cells were collected at the indicated time points. (Left) Total RNAs were separated on a 1% formamideagarose gel, and MFA2pG mRNA was detected by Northern blotting. (Right) The amount of MFA2pG mRNAs at each time point was measured. The half-lives (t1/2) of the mRNAs were derived from three independent experiments and are shown as the average and the standard error of the mean (SEM) in the left panel. (D) The deadenylation rates of MFA2pG mRNA poly(A) tails in the strains shown in C were analyzed by a polyacrylamide Northern blot analysis. (Left) The RNA samples prepared in C were separated on a 4% polyacrylamide–8 M urea gel. The RNA samples from 0-min time points were treated with RNaseH and oligo(dT) to indicate the position of the deadenylated mRNA. (Right) The sizes of the longest poly(A) tails were measured.
However, a previous study reported that pan2Δ does not show any defect in the rate of mRNA deadenylation or decay (Tucker et al. 2001). Therefore, we next examined the decay of an MFA2pG reporter transcript that was under the control of a tetracycline-regulatable promoter system. In this system, the reporter gene is specifically and rapidly suppressed in the presence of doxycycline. The decay rate of the mRNA was measured after transcriptional shutoff by the addition of doxycycline. Consistent with previous reports, pan2Δ did not show any apparent defect in the decay of the MFA2pG mRNA (t1/2 = 15 min, cf. wild type and pan2Δ) (Fig. 1C). In addition, the poly(A) tail of MFA2pG examined using high-resolution polyacrylamide Northern blot shortened at a rate of 2.0 nt per minute, which was comparable with the rate in wild-type cells (Fig. 1D). In sharp contrast, both the decay and the poly(A) tail-shortening rates of the MFA2pG mRNA were substantially reduced in the strain expressing SUP35N (t1/2 = 46 min and 0.45 nt per minute), although the distribution of the poly(A) tail length at time 0 was comparable with that of pan2Δ. Furthermore, overexpression of SUP35N reduced the rates of both the decay and the poly(A) tail shortening of the mRNA in the pan2Δ strain. These results suggest that additional deadenylases are involved in eRF3-mediated deadenylation and decay. In view of the defect in the deadenylation rates, Ccr4 is one such candidate. As reported previously, both the decay and the poly(A) tail shortening of the MFA2pG mRNA were substantially reduced in the strain ccr4Δ (t1/2 = 68 min and 0.35 nt per minute) (Tucker et al. 2001), which is similar to the phenotype of the SUP35N-overexpressing strain. In addition, SUP35N had no significant effect on the rates of both deadenylation and decay of the mRNA in the ccr4Δ strain but led to an increase in the poly(A) tail length of either MFA2 mRNA or bulk mRNA in the ccr4Δ strain at steady state (Fig. 1, cf. D and A). Thus, the phenotype of the longer poly(A) tail of steady-state mRNA in the SUP35N-overexpressing strain is explained by the defect in Pan2, and the phenotype of the reduced rates of both deadenylation and decay is explained by the defect in Ccr4. Taken together, these results indicate that two major mRNA deadenylases, Pan2–Pan3 and Caf1–Ccr4, are involved in the eRF3-mediated deadenylation of mRNA.
The two major mRNA deadenylases, hPan2–hPan3 and hCaf1–hCcr4, are involved in the eRF3-mediated deadenylation of mRNA in mammals
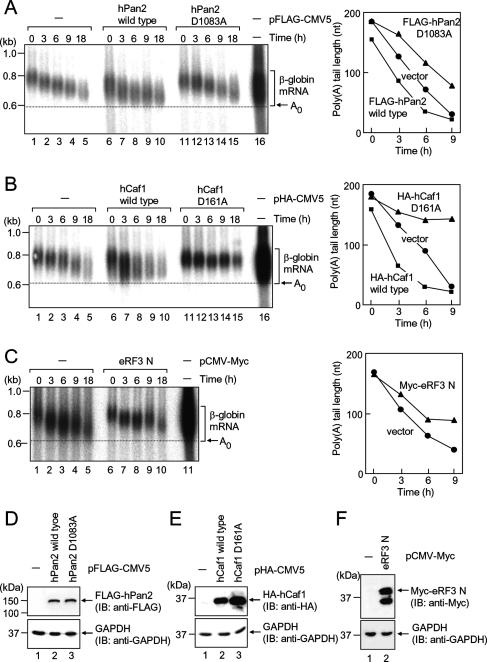
Next, we examined whether hPan2–hPan3 and hCaf1–hCcr4 are also involved in the eRF3-mediated mRNA deadenylation in mammalian cells. We monitored mRNA deadenylation and decay kinetics by using a tetracycline regulatory transcriptional pulse-chase approach in HeLa cells. In this experiment, β-globin mRNA was used as a reporter, since the transcript is well characterized and therefore provides an ideal model system to study mRNA stability. T-Rex-HeLa cells were transfected with the β-globin gene, transcription of the gene was driven by tetracycline for a short period of time, and the metabolism of newly transcribed mRNAs was followed over time. As reported previously, β-globin mRNA showed slow and biphasic decay kinetics with a half-life of ∼12 h (Fig. 2A, left panel). A slow and synchronous deadenylation occurred in the first phase (0–3 h), which was followed by a fast and less-synchronous deadenylation with a concomitant decay of the mRNA body (second phase) (Supplementary Fig. 1C, left panel). Overexpression of wild-type hPan2 increased the rate of deadenylation in the first phase (0–3 h) (Fig. 2A, lanes 6,7), whereas a comparable amount of hPan2 mutant that had no deadenylase activity (Uchida et al. 2004) reduced the rate (Fig. 2A, lanes 11,12). On the other hand, overexpression of wild-type hCaf1 increased the rate of deadenylation (Fig. 2B; Supplementary Fig. 1) and enhanced the decay of the mRNA body in the second phase (Fig. 2B, lane 7; Supplementary Fig. 1C). In sharp contrast to this, a nuclease-deficient hCaf1 mutant in which a catalytically essential aspartate residue was mutated to alanine drastically reduced the rate of deadenylation and decay of mRNA body in the second phase and stabilized the mRNA (Fig. 2B, lanes 13–15; Supplementary Fig. 1C). The expression of hPan2 or hCaf1 protein was confirmed by Western blot (Fig. 2D,E). These results confirm the previous results showing that hPan2–hPan3 and hCaf1–hCcr4 function in the first and second phases of mRNA deadenylation, respectively (Yamashita et al. 2005).
Figure 2.
Comparison of deadenylation rates in HeLa cells. (A) T-Rex-HeLa cells were transfected with pFlag-CMV5/TO-G1 reporter and either pFlag-CMV5 (lanes 1–5), pFlag-hPan2 (lanes 6–10), or pFlag-hPan2 D1085A (lanes 11–15). One day later, β-globin mRNA was induced by tetracycline for 2.5 h, and then cells were harvested at the specified time after β-globin mRNA transcription shutoff. (Left) β-Globin mRNA was detected by Northern blot analysis. (Lane 16) To determine the overall poly(A) tail length and the deadenylation endpoint (arrow) of β-globin mRNA, β-globin mRNAs at the steady-state level (24-h induction) are shown. (Right) The average length of poly(A) tails at each time point was measured. (B) As in A, except that cells were transfected with pHA-CMV5 (lanes 1–5), pHA-hCaf1 (lanes 6–10), or pHA-hCaf1 D161A (lanes 11–15), and transcriptional pulse-chase analysis was performed. (C) As in A, except that cells were transfected with pCMV-Myc (lanes 1–5) or pCMV-Myc-eRF3 N (lanes 6–10), and transcriptional pulse-chase analysis was performed. (D–F) Total cell lysates were analyzed by immunoblotting using anti-Flag (D, top), anti-HA (E, top), anti-Myc (F, top), or anti-GAPDH (D–F, bottom).
We then examined whether eRF3 mediates deadenylation of mRNA in HeLa cells. As shown in Figure 2C, ectopic expression of an N-terminal domain of eRF3 (eRF3N) reduced the rate of deadenylation in both the first and the second phases. The expression of the eRF3N was confirmed by Western blot (Fig. 2F). Collectively, these results suggest that eRF3 mediates deadenylation through hPan2–hPan3 and hCaf1–hCcr4 in mammalian cells.
hPan3 and eRF3 competitively bind to PABPC1
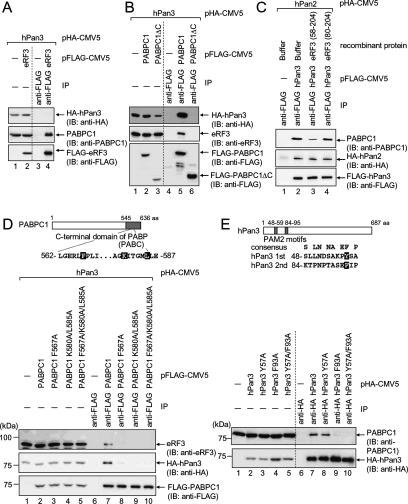
In mammals, we showed that eRF3 as well as hPan3 interacts with PABPC1 (Hoshino et al. 1999; Uchida et al. 2002, 2004). The present results showing that eRF3-mediated deadenylation is catalyzed in part by hPan2–hPan3 (Fig. 2) led us to speculate that, in mammalian cells, eRF3 mediates mRNA deadenylation by hPan2–hPan3 through interaction with PABPC1. Therefore, we investigated the interactions among eRF3, hPan2–hPan3, and PABPC1. To this end, we first examined whether eRF3 and hPan3 could simultaneously interact with PABPC1. When an extract of COS-7 cells expressing Flag-eRF3 and HA-hPan3 was immunoprecipitated with anti-Flag antibody, the precipitated fraction contained endogenous PABPC1 but not HA-hPan3 (Fig. 3A, lane 4). On the other hand, anti-Flag immunoprecipitate from extracts of cells expressing Flag-hPan3 contained PABPC1 but not eRF3 (data not shown). These results suggest that both hPan3 and eRF3 bind with PABPC1, but each protein is part of a distinct complex, and eRF3 might compete with hPan3 for PABPC1 binding. To test this idea, we examined whether hPan3 and eRF3 interact with the same region of PABPC1. Flag-PABPC1 or Flag-PABPC1ΔC lacking the eRF3-binding domain was coexpressed with HA-hPan3, and an immunoprecipitation assay using anti-Flag antibody was performed. As shown in Figure 3B, Flag-PABPC1, but not Flag-PABPC1ΔC was capable of coprecipitating with eRF3, which is consistent with our previous study using a yeast two-hybrid system (Hoshino et al. 1999). A similar result was also obtained for the coprecipitation of HA-hPan3 (Fig. 3B, top). Thus, both eRF3 and hPan3 require the C-terminal domain of PABPC1 for their binding.
Figure 3.
hPan3 and eRF3 competitively bind to PABC domain of PABPC1 via their PAM2 motifs. (A) COS-7 cells were transfected with pHA-hPan3 and either pFlag-CMV5 (lanes 1,3) or pFlag-eRF3 (lanes 2,4). The cell extracts were subjected to an immunoprecipitation assay (IP) using anti-Flag antibody. The immunoprecipitates (lanes 3,4) and the inputs (lanes 1,2, 20% of the amount immunoprecipitated) were analyzed by immunoblotting using the indicated antibodies. (B) COS-7 cells were transfected with pHA-hPan3 and either pFlag-CMV5 (lanes 1,4), pFlag-PABPC1 (lanes 2,5), or pFlag-PABPC1ΔC (lanes 3,6). The cell extracts were subjected to an immunoprecipitation assay and immunoblot analyses as described in A. (C) COS-7 cells were transfected with pHA-hPan2 and either pFlag-CMV5 (lane 1) or pFlag-hPan3 (lanes 2–4), and the cell extracts were subjected to an immunoprecipitation assay using anti-Flag antibody. Coprecipitated proteins were incubated with buffer alone (lane 2), recombinant GST-eRF3 (58–204)-(His)6 (lane 3), or GST-eRF3 (80–204)-(His)6 (lane 4). The remaining proteins were separated by SDS-PAGE, and immunoblot analysis was carried out with the indicated antibodies. (D) COS-7 cells were transfected with pHA-hPan3 and either pFlag-CMV5 (lanes 1,6), pFlag-PABPC1 (lanes 2,7), pFlag-PABPC1 F567A (lanes 3,8), pFlag-PABPC1 K580A/L585A (lanes 4,9), or pFlag-PABPC1 F567A/K580A/L585A (lanes 5,10). The cell extracts were subjected to an immunoprecipitation assay and immunoblot analysis as described in A. (Top) PABPC1 protein is schematically represented. Gray bar indicates PABC domain. The partial sequence of PABC is also shown. The amino acid residues that were mutated in the experiment are shaded. (E) COS-7 cells were transfected with either pHA-CMV5 (lanes 1,6), pHA-hPan3 (lanes 2,7), pHA-hPan3 Y57A (lanes 3,8), pHA-hPan3 F93A (lanes 4,9), or pHA-hPan3 Y57A/F93A (lanes 5,10). The cell extracts were subjected to an immunoprecipitation assay using anti-HA antibody. The immunoprecipitates (lanes 6–10) and the inputs (lanes 1–5, 10% of the amount immunoprecipitated) were analyzed by immunoblotting using the indicated antibodies. (Top) hPan3 protein is schematically represented. Gray bars indicate putative PAM2 motifs. The sequences of the two putative PAM2 motifs are also shown. The amino acid residues that were mutated in the experiment are shaded.
Next, we examined whether hPan3 actually competes with eRF3 for PABPC1 binding. A complex consisting of Flag-hPan3, HA-hPan2, and PABPC1 was prepared from COS-7 cells expressing both Flag-hPan3 and HA-hPan2 by means of immunoprecipitation with anti-Flag antibody. Then the complex was incubated with recombinant eRF3 (58–204) (a fragment encompassing amino acids 58–204) or eRF3 (80–204), which contained the PABPC1-binding region or not, respectively (Uchida et al. 2002). As shown in Figure 3C, eRF3 (58–204) but not eRF3 (80–204) effectively inhibited PABPC1 binding to Flag-hPan3, while the interaction between HA-hPan2 and Flag-hPan3 was not affected. Thus, we conclude that hPan3 competes with eRF3 for PABPC1 binding.
The PAM2 motif of hPan3 and PABC domain of PABPC1 are responsible for the interaction between hPan3 and PABPC1
We reported previously that a 22-amino-acid region at the N terminus of eRF3 is critical for the binding to the C-terminal domain of PABPC1 (PABC) (Uchida et al. 2002). The PABC domain has been characterized as a protein–protein interaction domain that recruits various translation or mRNA-processing factors possessing the PABPC1-interacting motif (PAM2) to the mRNA poly(A) tail (Kozlov et al. 2001). The PAM2 motif is also found in the N-terminal 22-amino-acid region of eRF3 (Uchida et al. 2002). Thus, eRF3 binds to PABC through its PAM2 motif. hPan3 also binds to the C-terminal domain of PABPC1 competitively with eRF3. These results led us to speculate that hPan3 competes with eRF3 by binding to the PABC domain of PABPC1. As expected, we identified two putative PAM2 motifs at the N-terminal region of hPan3 (Fig. 3E). We therefore tested the idea by site-directed mutagenesis of the PABC domain and immunoprecipitation with extracts from COS-7 cells expressing the mutants. As shown in Figure 3D, the binding of hPan3 with PABPC1 was abolished by mutation of Phe567 to alanine (F567A) (lane 8), and also by a double (K580A/L585A) (lane 9) and a triple (F567A/K580A/L585A) (lane 10) mutation in the PABC domain of PABPC1. The same results were obtained for eRF3 (Fig. 3D, top panel). These results indicate that hPan3 as well as eRF3 binds to PABC. Next, we examined whether hPan3 binds to PABC domain through its PAM2 motifs. As shown in Figure 3E, the binding of PABPC1 was not affected by mutation of Tyr57 to alanine (Y57A) (lane 8) in the first PAM2 motif of hPan3, but was almost completely abolished by F93A mutation in the second PAM2 motif (lane 9) and also by their double mutation (Y57A/F93A) (lane 10). Thus, hPan3 binds to PABPC1 with the second PAM2 motif. From these results, we conclude that hPan3 and eRF3 competitively bind to the PABC domain of PABPC1 through their PAM2 motifs.
Caf1–Ccr4 interacts with PABPC1 through Tob by using PAM2–PABC contact
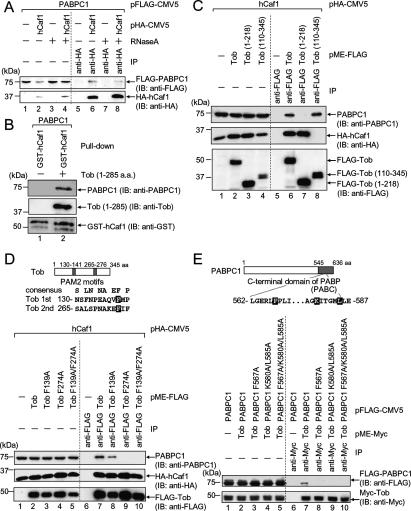
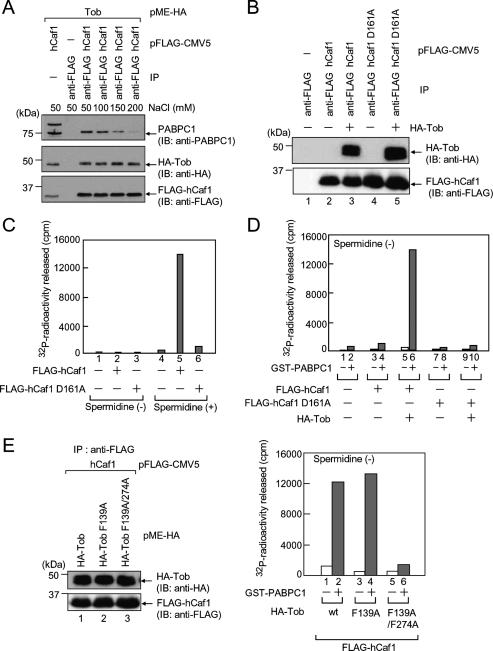
In a previous study, we demonstrated that PABPC1 is required for the activation of the deadenylating activity of hPan2–hPan3 in vitro (Uchida et al. 2004). However, the role of PABPC1 in the activity of the hCaf1–hCcr4 complex is still controversial; in yeast, Pab1 is inhibitory to Ccr4 in vitro (Tucker et al. 2002), whereas a pab1Δ strain severely blocks Ccr4-catalyzed deadenylation in vivo (Caponigro and Parker 1995). To address whether PABPC1 is also involved in the activation of the hCaf1–hCcr4 complex, we first examined the physical interaction between hCaf1 and PABPC1 by an immunoprecipitation experiment. HeLa cells expressing Flag-PABPC1 and HA-Caf1 were immunoprecipitated using anti-HA antibody. Flag-PABPC1 was capable of coprecipitating with HA-hCaf1 (Fig. 4A, lane 6). RNaseA treatment did not abolish the binding, which indicates that no RNA was required for the binding (Fig. 4A, lane 8). Moreover, the binding is indirect because a GST pull-down experiment using purified recombinant PABPC1 and GST-hCaf1 showed no interaction (Fig. 4B, lane 1). We therefore searched for a factor that mediates hCaf1–PABPC1 binding. Previous findings independently demonstrated that Tob binds to both PABPC1 (Okochi et al. 2005) and hCaf1 (Ikematsu et al. 1999). This led us to speculate that Tob mediates hCaf1–PABPC1 binding. As expected, purified recombinant PABPC1 bound to GST-hCaf1 in the presence of Tob (Fig. 4B). To further confirm that Tob acts as a bridge between hCaf1 and PABPC1, we examined the structural requirements for the interaction. Either Flag-tagged Tob or its deletion mutants were expressed in COS-7 cells, and immunoprecipitation experiments using anti-Flag antibody were performed. Flag-Tob coprecipitated with both PABPC1 and HA-Caf1 (Fig. 4C, lane 6). On the other hand, truncation of the Tob C terminus (127 amino acids) led to a loss of interaction with PABPC1 but not with HA-hCaf1. Conversely, N-terminal truncation of Tob (109 amino acids) led to a loss of interaction with HA-hCaf1 but not with PABPC1. Taken together, these results indicate that PABPC1 and hCaf1 bind to the C- and N-terminal region of Tob, respectively, to form a complex.
Figure 4.
hCaf1 binds to PABC domain of PABPC1 via Tob with PAM2 motifs. (A) HeLa cells were transfected with pFlag-PABPC1 and either pHA-CMV5 (lanes 1,3,5,7) or pHA-hCaf1 (lanes 2,4,6,8). The cell extracts were subjected to an immunoprecipitation assay using anti-HA antibody in the presence or absence of RNaseA. The immunoprecipitates (lanes 5–8) and the inputs (lanes 1–4, 10% of the amount immunoprecipitated) were analyzed by immunoblotting using the indicated antibodies. (B) Recombinant PABPC1 and GST-hCaf1 prepared from E. coli were subjected to a GST pull-down assay in the presence (lane 2) or absence (lane 1) of recombinant Tob (1–285). Immunoblot analyses were performed with the indicated antibodies. (C) COS-7 cells were transfected with pHA-hCaf1 and either pMEFlag (lanes 1,5), pMEFlag-Tob (lanes 2,6), pMEFlag-Tob (1–218) (lanes 3,7), or pMEFlag-Tob (110–345) (lanes 4,8). The cell extracts were subjected to an immunoprecipitation assay using anti-Flag antibody. The immunoprecipitates (lanes 5–8) and the inputs (lanes 1–4, 20% of the amount immunoprecipitated) were analyzed by immunoblotting using the indicated antibodies. (D) COS-7 cells were transfected with pHA-hCaf1 and either pMEFlag (lanes 1,6), pMEFlag-Tob (lanes 2,7), pMEFlag-Tob F139A (lanes 3,8), pMEFlag-Tob F274A (lanes 4,9), or pMEFlag-Tob F139A/F274A (lanes 5,10). The cell extracts were subjected to an immunoprecipitation assay and immunoblot analyses as described in C. (Top) Tob protein is schematically represented. Gray bars indicate putative PAM2 motifs. The sequences of the two putative PAM2 motifs are also shown. The amino acid residues that were mutated in the experiment are shaded. (E) COS-7 cells were transfected with pMEMyc- Tob and either pFlag-PABPC1 (lanes 2,7), pFlag--PABPC1 F567A (lanes 3,8), pFlag-PABPC1 K580A/L585A (lanes 4,9), or pFlag-PABPC1 F567A/K580A/L585A (lane 5,10). (Lanes 1,6) As a control, COS-7 cells were transfected with pMEMyc and pFlag-PABPC1. The cell extracts were subjected to an immunoprecipitation assay using anti-Myc antibody. The immunoprecipitates (lanes 6–10) and the inputs (lanes 1–5, 5% of the amount immunoprecipitated) were analyzed by immunoblotting using the indicated antibodies.
The above results demonstrate that a PAM2 motif of hPan3 and the PABC domain of PABPC1 play essential roles for their binding. We therefore asked whether the combination of PAM2 and PABC is also involved in the interaction between Tob and PABPC1 first by searching for PAM2 motifs in the sequence of Tob. As expected, we identified two putative PAM2 motifs in the C-terminal region of Tob (Fig. 4D). Recently, PAM2 motifs in Tob2, a distinct subtype of Tob, have been reported (Lim et al. 2006). We therefore approached the above question by site-directed mutagenesis and immunoprecipitation with extracts from COS-7 cells expressing the mutants. As shown in Figure 4D, mutation of Phe139 to alanine (F139A) in the first PAM2 motif had no effect on the binding of PABPC1, whereas mutation of Phe274 to alanine (F274A) in the second PAM2 motif completely abolished the binding. These results indicate that PABPC1 binds to the second PAM2 motif of Tob. Next we examined whether Tob binds to the PABC domain of PABPC1. As shown in Figure 4E, the binding of Tob was completely abolished by mutation of Phe567 to alanine (F567A) (lane 8), and also by a double (K580A/L585A) (lane 9) and a triple (F567A/K580A/L585A) mutation (lane 10) in PABPC1. Thus, we conclude that Tob binds to the PABC domain of PABPC1 through a PAM2 motif and bridges between PABPC1 and Caf1 to form a complex.
A translation-dependent exchange of eRF3 for hPan2–hPan3 occurs on PABPC1 in living cells
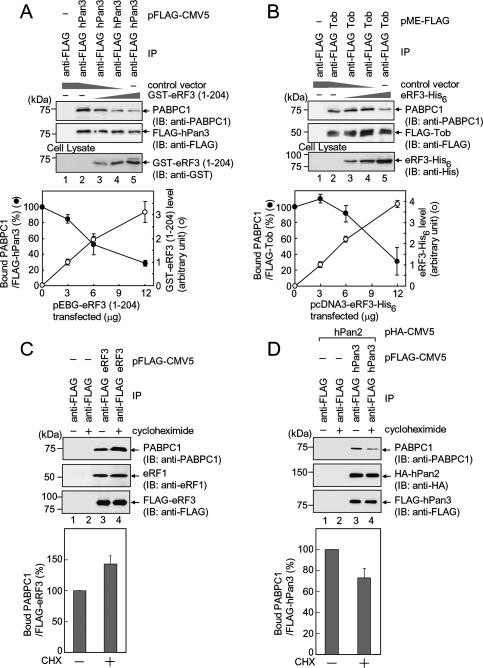
Based on the structural evidence described above, we next examined the possibility that competition among eRF3, hCaf1–hCcr4, and hPan2–hPan3 occurs on PABPC1 in living cells. First, Flag-hPan3 and GST-eRF3 (1–204) were expressed in COS-7 cells, and immunoprecipitation experiments were performed with anti- Flag antibody. The amount of PABPC1 coprecipitated with Flag-hPan3 was decreased by increasing the amount of GST-eRF3 expressed in cells (Fig. 5A, top). Similar results were obtained with Tob; the amount of PABPC1 coprecipitated with Flag-Tob was inversely correlated with the amount of eRF3-(His)6 expressed in cells (Fig. 5B, top). These results suggest the possibility that hPan2–hPan3 and hCaf1–hCcr4 each competes with eRF3 for the binding with PABPC1 in living cells.
Figure 5.
Both hPan3 and Tob compete with eRF3 to form a complex with PABPC1 in a translation-dependent manner. (A) COS-7 cells were transfected with pFlag-hPan3 and increasing amounts of pEBG-eRF3 (1–204) (0, 3, 6, and 12 μg in lane 2, 3, 4, and 5, respectively). The total amounts of plasmid introduced into the cells were maintained at the same level by cotransfecting the control vector. The cell extracts were subjected to an immunoprecipitation assay using anti-Flag antibody. (Top) Immunoblot analyses were performed with the indicated antibodies. (Bottom) The ratios of bound PABPC1 to precipitated Flag-hPan3 and the expression level of GST-eRF3 (1–204) in the cell lysates were measured. Error bars indicate SEM of three independent experiments. (B, top) COS-7 cells were transfected with pMEFlag-Tob and increasing amounts of pcDNA3-eRF3-(His)6, and an immunoprecipitaion assay was performed as described in A. (Bottom) The ratios of bound PABPC1 to precipitated Flag-Tob and the expression level of eRF3-(His)6 in the cell lysates were measured. Error bars indicate SEM of three independent experiments. (C) COS-7 cells were transfected with pFlag-CMV5 or pFlag-eRF3 and treated with or without cycloheximide (100 μg/mL) for 4 h. The cell extracts were subjected to immunoprecipitation assay using anti-Flag antibody. (Top) Immunoblot analyses were performed with the indicated antibodies. (Bottom) The ratios of bound PABPC1 to precipitated Flag-eRF3 were calculated. Error bars indicate SEM of at least four independent experiments. (D) COS-7 cells were transfected with pHA-hPan2 and either pFlag-CMV5 or pFlag-hPan3, and CHX treatment and immunoprecipitation assay were performed as described in C.
Since hPan2–hPan3 and hCaf1–hCcr4 are involved in eRF3-mediated mRNA deadenylation, which is coupled to translation termination, we expected that a change of translation status might affect interactions of eRF3, hPan3, and Tob with PABPC1. Therefore, we examined the effect of a translation inhibitor, cycloheximide (CHX), on the interactions. Cells expressing Flag-eRF3 were treated with CHX and subjected to immunoprecipitation using anti-Flag antibody. Figure 5C shows that the amounts of Flag-eRF3 and eRF1 precipitated were almost the same in the presence and absence of CHX, while CHX treatment markedly increased the amount of coprecipitated PABPC1. In contrast, a comparative experiment conducted using cells expressing Flag-hPan3 and HA-Pan2 showed that the CHX treatment remarkably attenuated the interaction between PABPC1 and hPan3 without affecting that between hPan3 and hPan2 (Fig. 5D). Thus, CHX treatment had opposite effects on eRF3–PABPC1 and hPan3–PABPC1 interactions. These results suggest that a translation-dependent exchange of eRF3 and hPan3 occurs on PABPC1. Tob was refractory to this analysis, since Tob has a quick turnover through the ubiquitin–proteasome pathway and CHX treatment greatly affects the amount of Tob.
Deadenylating activities of both hCaf1–hCcr4 and hPan2–hPan3 are stimulated by PABPC1
We demonstrated previously that the binding of PABPC1 to hPan2–hPan3 leads to the activation of the deadenylase (Uchida et al. 2004). In order to assess the generality of this observation for hCaf1–hCcr4, we tested the effect of PABPC1 on the activity of hCaf1. We first prepared the hCaf1–Tob complex. Flag-tagged hCaf1 and HA-tagged Tob were coexpressed in COS-7 cells, and the cell extracts were subjected to immunoprecipitation with anti-Flag IgG beads. To deplete the copurified PABPC1 from the hCaf1–Tob complex, the precipitated beads were washed with buffer containing 200 mM NaCl. This washing markedly attenuated the interaction between Tob and PABPC1 without affecting the interaction between hCaf1 and Tob (Fig. 6A). Thus, >85% of PABPC1 could be removed from the immunopurified preparation as compared with that prepared without the high-salt wash. As shown in Figure 6B, Tob was copurified with hCaf1 as a complex (Tob–hCaf1 complex). For comparison, we also prepared a nuclease-deficient hCaf1 mutant (D161A) as a complex with Tob (Tob–D161A complex). The RNase activities of the purified proteins were examined in the presence or absence of spermidine. Figure 6C shows that the purified Tob–hCaf1 complex could degrade poly(A) RNA in the presence of spermidine (column 5), whereas the Tob–D161A complex had no RNase activity (column 6). As in the case of hPan2–hPan3, none of the proteins exhibited any detectable RNase activity in the absence of spermidine (Fig. 6C, columns 1–3). Under this assay condition, the effect of PABPC1 on RNase activity was assessed. As shown in Figure 6D, the recombinant GST-fused PABPC1 prepared from Escherichia coli could markedly stimulate the RNase activity of the Tob–hCaf1 complex (column 6). In sharp contrast, GST-PABPC1 could not stimulate the RNase activity of the Tob–D161A complex (Fig. 6D, column 10) or hCaf1 not complexed with Tob (Fig. 6D, column 4). These results indicate that PABPC1 stimulates hCaf1 through its association with Tob, consistent with the above finding that Tob mediates hCaf1 binding to PABPC1. To further confirm the significance of Tob in the PABPC1-stimulated hCaf1 activity, we also prepared hCaf1 in a complex with Tob whose PAM2 motifs were mutated. As shown in Figure 6E, F139A mutation of the first PAM2 motif of Tob had no significant effect on the PABPC1-stimulated RNase activity (column 4), whereas F139A/F274A mutations, which abrogated the Tob–PABPC1 binding, almost abolished the effect of PABPC1. Taken together with previous findings on hPan2–hPan3, these results suggest that recruitment of hPan2–hPan3 and hCaf1–hCcr4 to the poly(A)-bound PABPC1 is the general mechanism leading to the activation of the mRNA deadenylases.
Figure 6.
Tob-mediated binding of PABPC1 leads to the activation of hCaf1. (A) COS-7 cells were transfected with pFlag-hCaf1 and pME-HA–Tob. The cell extracts were subjected to immunoprecipitation using anti-Flag IgG agarose. After washing with buffer containing the indicated concentrations of NaCl, proteins were eluted with the Flag peptide from the immunoprecipitated beads. The precipitated proteins were immunoblotted with the indicated antibodies. (B) COS-7 cells were transfected with pFlag-hCaf1 (lanes 2,3) or pFlag-hCaf1 D161A (lanes 4,5) and either pMEFlag (lanes 2,4) or pMEFlag-Tob (lanes 3,5). The cell extracts were subjected to immunoprecipitation as described in A. Buffer containing 200 mM NaCl was used for washing the beads. (C) The purified Flag-tagged hCaf1 and hCaf1 (D161A) (as shown in B), were incubated with internally 32P-radiolabeled poly(A) RNA for 30 min at 30°C in the presence or absence of 1 mM spermidine, and 32P radioactivity released was measured. (D) The purified proteins (as shown in B) were incubated with the 32P-radiolabeled poly(A) RNA for 30 min at 30°C with or without 10 ng of GST-fused PABPC1 in the absence of spermidine. (E, left) COS-7 cells were transfected with pFlag-hCaf1 and either pMEHA–Tob (lane 1), pMEHA–Tob (F139A) (lane 2), or pMEHA–Tob (F139A/F274A) (lane 3). The cell extracts were subjected to immunoprecipitation as described in B. (Right) The purified proteins were incubated with the 32P-radiolabeled poly(A) RNA as described in D.
Significant roles of PABPC1-binding PAM2 motifs of Tob, hPan3, and eRF3 for mRNA deadenylation in cells
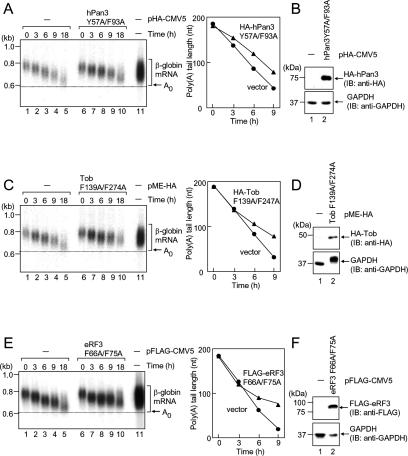
The above results strongly suggest that PAM2 motifs of Tob and hPan3 play essential roles in mRNA deadenylation by recruiting hCaf1–hCcr4 and hPan2 to poly(A)-bound PABPC1. To test this possibility, we applied a dominant-negative approach. T-Rex-HeLa cells were transfected with a β-globin reporter plasmid, and the deadenylation and decay kinetics were examined by transcriptional pulse-chase as described in Figure 2. As shown in Figure 7A, ectopic expression of hPan3 with PAM2 mutations (Y57A/F93A) slowed the rate of deadenylation in the first phase compared with the control case. The kinetics were similar to those seen with the hPan2 D1083A mutant (Fig. 2A). In sharp contrast, ectopic expression of Tob with PAM2 mutations (F139A/F247A) reduced the rate of deadenylation in the second phase without affecting the rate in the first phase (Fig. 7C), with kinetics resembling those seen with the hCaf1 D161A mutant (Fig. 2B). These results provide evidence that Tob actually functions with hCaf1–hCcr4 in the second phase of mRNA deadenylation, and further support the notion that PAM2 motifs play essential roles in triggering mRNA deadenylation. We also examined the effect of PAM2 mutations in eRF3 (F66A/F75A), which abrogated the binding to PABPC1 (Supplementary Fig. 3). Ectopic expression of the mutant slowed the rate of deadenylation mainly in the second phase (Fig. 7E). These results indicate that PAM2 motifs of Tob, hPan3, and eRF3—namely, PABPC1 binding—play key roles in eRF3-mediated mRNA deadenylation.
Figure 7.
hPan3, Tob, and eRF3 with mutations in the PABPC1-binding PAM2 motifs affect mRNA deadenylation in a dominant-negative fashion. (A) T-Rex-HeLa cells were transfected with pFlag-CMV5/TO-G1 reporter and either pHA-CMV5 (lanes 1–5) or pHA-hPan3 Y57A/F93A (lanes 6–10). Transcriptional pulse-chase analysis was performed as described in Figure 2. (B) Total cell lysates were analyzed by immunoblotting using anti-HA (top) and anti-GAPDH (bottom). (C) As in A, except that the cells were transfected with pMEHA or pMEHA–Tob F139A/F274A, and transcriptional pulse-chase analysis was performed. (D) As in B, total cell lysates were immunoblotted. (E) As in A, except that the cells were transfected with pFlag-CMV5 or pFlag-eRF3 F66A/F75A, and transcriptional pulse-chase analysis was performed. (F) As in B, except that anti-Flag antibody was used, and total cell lysates were immunoblotted.
Discussion
We demonstrated previously that eRF3 mediates deadenylation and decay of normal mRNA in a manner coupled to translation termination. In this study, we clarified the underlying mechanism of the eRF3-mediated mRNA deadenylation. The main findings of this study are (1) eRF3 mediates deadenylation through Pan2–Pan3 and Caf1–Ccr4 deadenylases, which are conserved from yeast to humans (Figs. 1, 2); (2) the termination complex eRF1–eRF3 and deadenylase complexes hPan2–hPan3 and hCaf1–hCcr4 interact with PABPC1, in all cases via PAM2–PABC contacts (Figs. 3, 4); (3) hPan2–hPan3 binds PABPC1 competitively with eRF3 and in a manner dependent on translation (Fig. 5); and (4) the binding of hPan2–hPan3 and hCaf1–hCcr4 with PABPC1 leads to the activation of the deadenylase activity (Fig. 6; Uchida et al. 2004). Together, these new findings define PABPC1 as a regulatory platform for mRNA deadenylation, and argue that the recruitment of deadenylating enzymes to PABPC1 is involved in the mechanism of activation of mRNA deadenylation. The data suggest that eRF3 mediates mRNA deadenylation by triggering the recruitment of the deadenylase complexes to PABPC1; as translation proceeds, the termination complex is released from PABPC1 and in turn the deadenylase complexes bind to PABPC1, which leads to the activation of the deadenylating enzymes.
General mechanism of mRNA deadenylation
Several previous reports using specific mRNAs showed that recruitment of deadenylating enzymes to the mRNA is involved in the mechanism of mRNA deadenylation. First, in Drosophila embryos, an RNA-binding protein, Smaug, which physically interacts with the Caf1–Ccr4 complex, recruits the complex to Hsp83 mRNA to trigger deadenylation and decay of the transcript. The recruitment-induced enzyme activation was confirmed by tethering Smaug to reporter transcripts (Semotok et al. 2005). Second, a similar tethering strategy was used to show that selective recruitment of Caf1–Ccr4 is sufficient to activate mRNA decay in yeast (Finoux and Seraphin 2006). Third, yeast Mpt5, a member of the PUF family of RNA-binding proteins, also recruits the Caf1–Ccr4 complex to HO mRNA to trigger deadenylation of the message (Goldstrohm et al. 2006). Fourth, an RNA-binding protein, CUG-BP, specifically binds to ARE-containing mRNAs to stimulate poly(A) shortening by recruiting another poly(A)-specific ribonuclease, PARN, to the mRNA (Moraes et al. 2006). For the hPan2–hPan3 deadenylase complex, we demonstrated previously that hPan3 recruits the hPan2 catalytic subunit to poly(A)-bound PABPC1 to activate its poly(A) nuclease activity (Uchida et al. 2004), which is in accord with a previous report showing that Pan2–Pan3 is a Pab1-dependent poly(A) nuclease in yeast (Brown et al. 1996). In this study, we further demonstrated that hCaf1–hCcr4 is also activated by its recruitment to poly(A)-bound PABPC1.
Our results indicate that anti-proliferative protein Tob is a key component of hCaf1–hCcr4. We presented three important observations showing physical and functional interaction between hCaf1–hCcr4 and PABPC1, which is supported by structural evidence. First, hCaf1 indirectly interacts with PABPC1 through Tob (Fig. 4A,B). Second, the binding of PABPC1 leads to the stimulation of hCaf1 deadenylase (Fig. 6). Third, the binding requires the PAM2 motif of Tob and the PABC domain of PABPC1 (Fig. 4D,E). The PAM2–PABC contact is also utilized for the interaction between hPan3 and PABPC1 (Fig. 3D,E). Thus, recruitment of both of the two major mRNA deadenylase complexes to poly(A)-bound PABPC1 through adaptor proteins is an important determinant in the activation of the deadenylases. These results suggest the biological significance of the mechanism of activation of mRNA deadenylases by PABPC1 in the regulation of mRNA deadenylation. One possible role of the adaptor proteins (Tob and hPan3) is to recruit mRNA deadenylase specifically to the substrate mRNA poly(A) tails by taking advantage of the poly(A) specificity of PABPC1. In the case of hPAN, the adaptor protein hPan3-mediated PABPC1 interaction actually increases its substrate specificity for poly(A) RNA (Uchida et al. 2004). The second possible role is to reduce entropy in the reaction by placing the deadenylase in close proximity to the substrate RNA. In the case of both deadenylases, >10-fold stimulation of the enzyme activity was observed when complexed with PABPC1 through the adapter.
The role of translation termination factor eRF3 in mRNA deadenylation
Our previous and present results show that eRF3 mediates deadenylation and decay of normal mRNA in a manner coupled to translation termination and the eRF3-mediated deadenylation is catalyzed by both hCaf1–hCcr4 and hPan2–hPan3. As translation proceeds, eRF3 is released from PABPC1, and hPan2–hPan3 (and possibly hCaf1–hCcr4) associates with PABPC1, which leads to the activation of the deadenylases. Thus, eRF3 triggers deadenylation by recruiting the deadenylases to poly(A)-bound PABPC1. Since eRF3 competes with the deadenylases for PABPC1 binding, dissociation of eRF3 from PABPC1 is likely to be the key determinant in triggering deadenylation. As in the case for other G protein family members, eRF3 is thought to function as a molecular switch that changes binding partners, depending on whether it is in its GTP- or GDP-bound form. Since ribosome and eRF1 stimulate GTP hydrolysis of eRF3 (Frolova et al. 1996), a conformational change of eRF3 during or after translation termination might trigger dissociation of eRF3 from PABPC1. Consistent with this idea, recent findings suggested that binding of PABPC1 and eRF1 to eRF3 might be mutually exclusive (Hauryliuk et al. 2006).
Our present study has focused on the role of eRF3 in normal mRNA decay. However, eRF3 also functions in NMD. NMD is a surveillance mechanism that degrades mRNA containing premature termination codons (PTCs). Since PTCs are termination codons, recognition of PTCs is thought to be mediated by translation termination. Recent findings demonstrated that eRF1–eRF3 binds the NMD factors SMG1–Upf1 to form a complex termed SURF, and an association between SURF and the exon junction complex triggers phosphorylation of Upf1 (Kashima et al. 2006). Although it is not known how Upf1 phosphorylation leads to rapid decay of PTC-containing mRNA, eRF3 must play a pivotal role in triggering NMD.
We and others showed previously that a member of the eRF3 family of G proteins, Ski7, interacts with the exosome to function in 3′-to-5′ decay of mRNA (van Hoof et al. 2000; Araki et al. 2001), and later, Ski7 was shown to be involved in the decay of aberrant mRNAs lacking termination codons, which is called “nonstop mRNA decay” (NSD) (Frischmeyer et al. 2002; van Hoof et al. 2002). In NSD, Ski7 is suggested to bind to a ribosome stalled at the 3′ end of the mRNA and recruit the exosome to the mRNA to trigger 3′-to-5′ decay of the mRNA. In addition, Hbs1, the closest relative of eRF3, has similar roles in another type of aberrant mRNA. mRNAs with ribosomes stalled before translation termination are recognized and degraded by a pathway referred to as “no-go decay” (NGD) (Doma and Parker 2006). In NGD, Hbs1 is suggested to recognize the stalled ribosome to trigger endonucleolytic cleavage of the mRNA. Thus, eRF3 family members play key roles in triggering mRNA decay by functioning as a molecular switch connecting translation to mRNA decay.
Future challenges include clarifying the mechanism of sequential deadenylation by hPan2–hPan3 and hCaf1–hCcr4 during poly(A) tail shortening of mRNA. As reported previously, our results also show that the poly(A) tail is first shortened in a distributive manner by hPan2–hPan3 to ∼100 nt (first phase, 0–3 h), and the remaining poly(A) tail is then rapidly and processively degraded by hCaf1–hCcr4 (Figs. 2, 7; Yamashita et al. 2005). It is not clear, however, why eRF3 triggers activation of hPan2–hPan3 in the first phase, but hCaf1–hCcr4 in the second phase. Recent work by Keeling et al. (2006) may provide a clue to this question. They found a novel factor (termed Tpa1) that influences translation termination and mRNA deadenylation in S. cerevisiae. Tpa1 interacts with both eRF1–eRF3 and Pab1, and is suggested to be involved in the termination-coupled mRNA deadenylation (Keeling et al. 2006). Interestingly, Tpa1 serves as a positive regulator of Pan2–Pan3 and a negative regulator of Caf1–Ccr4. Thus, Tpa1, through the interaction with eRF3, might positively and negatively regulate the recruitment of Pan2–Pan3 and Caf1–Ccr4 to Pab1, respectively (Keeling et al. 2006). Further studies will be required to test this possibility.
Note
During preparation of our manuscript, there appeared a paper (Siddiqui et al. 2007) in which the authors report a physicochemical analysis of PAM2–PABC contact between hPan3 and PABPC1 by using NMR. They showed that the interaction is conserved from yeast to humans, and is involved in the first trimming of mRNA poly(A) tails in yeast.
Materials and methods
Plasmids
To construct p9L-MFA2pG with the MFA2pG gene under the control of the tetO promoter, the cassette harboring the tetO promoter and CYC1 terminator was excised from pCM190 and inserted into YEplac181 to give p9L. MFA2pG fragment was excised from pGAL-MFA2pG (Araki et al. 2001) and subcloned into p9L. To construct pEBG-eRF3 (1–204) and pCMV-Myc-eRF3 N (1–204), the corresponding cDNA fragments of eRF3 were inserted in pEBG and pCMV-Myc (Clontech), respectively. In this study, we used the GSPT2 gene for the plasmids expressing eRF3. To construct pFlag-PABPΔC, a PCR fragment encoding the N-terminal 375 amino acids of PABPC1 was inserted in pFlag-CMV5 (Eastman Kodak Co.). To construct pMEFlag-Tob and pFlag-Caf1, the full-length Tob and hCAF1 cDNAs were amplified by RT–PCR using total RNA from HeLa cells and inserted into pMEFlag (a Flag-tagged vector based on pME18SFL3) and pFlag-CMV5, respectively. pMEHA-Tob and pHA-CAF1, which contain the HA tag instead of Flag, were also constructed. To generate pMEFlag-Tob (1–218) and pMEFlag-Tob (110–345), the corresponding cDNA fragments of Tob were amplified by PCR and inserted into pMEFlag. Point mutants in hCaf1, PABPC1, hPan3, Tob, and eRF3 were generated from pHA-hCaf1, pFlag-PABPC1, pHA-hPan3, pMEFlag-Tob and pFlag-eRF3, respectively, using a QuickChange Site-Directed Mutagenesis Kit and were confirmed by sequencing. pFlag-CMV5/TO-G1 was generated from pFlag-CMV5 by inserting the two Tet-operators and the β-globin gene downstream from its CMV promoter. Construction of all other plasmids used in this study has been described previously (Hosoda et al. 2003; Uchida et al. 2004).
Yeast strains and growth conditions
The yeast strains used were derived from MBS (MATa ade2 his3 leu2 trp1 ura3 can1) and are listed in Supplementary Table 1. Strains were grown in standard media. Disruption of yeast genes was achieved using standard PCR-based methods and the disruption was confirmed by PCR reactions.
Cell culture and DNA transfection
COS-7, HEK-293T, and T-REx-HeLa cells (Invitrogen) were cultured in Dulbecco’s modified Eagle’s medium (Nissui) supplemented with 10% fetal calf serum. Cells were transfected with plasmid using Lipofectin or LipofectAMINE 2000 (Invitrogen).
Immunoprecipitation
The transfected cells were lysed with 10 μg/mL of boiled RNaseA in buffer A consisting of 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1% Nonidet P-40, 1 mM dithiothreitol (DTT), 2.5 mM EDTA-Na (pH 8.0), 100 μM PMSF, 2 μg/mL of aprotinin, and 2 μg/mL of leupeptin. After centrifugation at 15,000g for 20 min, the lysate was incubated for 1 h at 4°C with anti-Flag IgG agarose (Sigma) or Anti-HA Affinity Matrix (Roche Diagnostics), and then the resin was washed with buffer A. As needed, recombinant proteins were added and the resin was further incubated for 60 min at 4°C. After washing with buffer A, proteins retained on the resin were subjected to SDS-PAGE and immunoblot analysis. The antibodies used for immunoblotting were anti-Flag M2 (Sigma), anti-HA (3F10, Roche Diagnostics), anti-Myc (9E10 [Sigma] or A-14 [Santa Cruz Biotechnology]), anti-GST (Santa Cruz Biotechnology), anti-His (MBL), anti-Tob (clone 4B1, Sigma), anti-PABP (kind gift from H. Imataka and N. Sonenberg), and anti-GSPT (Hoshino et al. 1998).
RNA analysis
For the Tet-off transcriptional repression experiment, yeast cells were grown in selective medium, which contained 2% galactose to produce the Sup35 N domain, for 6 h at 30°C (yeast cells reached mid-log phase [OD600 = 0.5–0.6] in this condition). Cells were collected by centrifugation and resuspended in 10 mL of selective media. After the cells were incubated for 10 min at 30°C, doxycycline was added to a final concentration of 5 μg/mL to repress transcription. At the indicated times, cells were harvested and immediately frozen in liquid nitrogen. For preparation of total RNA in the steady state, yeast cells were grown to mid-log phase (0.5–0.6) and collected by centrifugation. RNA extraction from the cell pellet was performed as described previously (Hosoda et al. 2003). To determine the poly(A) tail length of MFA2pG mRNA, total RNA was separated on a 4% polyacrylamide/8 M urea gel and electroblotted to Hybond XL (Amersham Pharmacia). For the analysis of mRNA decay, total RNA was separated on a 1% agarose gel and blotted to Hybond XL as described previously. The reporters were detected by Northern blotting using an end-labeled oligonucleotide, oRP121 (5′-AATTCCCCCCCCCCCCCCCCCCA-3′), which is complementary to the poly(G) sequence at the 3′-untranslated region of the transcript, as a probe. The poly(A) tail length analysis for total RNA was performed as described previously (Brown et al. 1996).
For the transcriptional pulse-chase analysis, T-Rex-HeLa cells were transfected with pFlag-CMV5/TO-G1. After 24 h, cells were treated with 10 ng/mL tetracycline for 2.5 h to induce transcription, and were harvested at the specified time after transcription shutoff. Total RNA was isolated with TriPure Reagents (Roche) and separated by 2.2% agarose gel electrophoresis using a NorthernMax Kit (Ambion). β-Globin mRNA was detected by Northern blotting using [32P]-labeled β-globin ORF as a probe.
In vitro nuclease assay
The production of internally 32P-radiolabeled poly(A) RNA was described previously (Uchida et al. 2004). In brief, oligo(A)23 was extended using [α-32P]ATP and yeast poly(A) polymerase (USB Corp.). The labeled poly(A) RNA was purified by gel filtration with S-400 HR MicroSpin columns (Amersham Bioscience) and ethanol precipitation. For the immunopurification of recombinant proteins, extracts prepared from COS-7 cells transfected with pFlag-hCAF1 and with either pMEHA or pMEHA-Tob were immunoprecipitated with anti-Flag antibody in buffer A. After washing three times with buffer A, the precipitated proteins were eluted with buffer B (10 mM HEPES-Na at pH 7.5, 2 mM MgCl2, 1 mM DTT, 0.02% Nonidet P-40) containing 100 μg/mL Flag peptides (Sigma). The immunopurified proteins were incubated for 30 min at 30°C in buffer B with 1 U/μL RNasin (Promega) and 1 × 105 counts per minute (cpm) of labeled poly(A) RNA in the presence or absence of 1 mM spermidine. If necessary, recombinant GST-fused PABPC1 was added to the reactions. The extent of nucleolysis was measured as follows: Aliquots were precipitated with 25% trichloroacetic acid by centrifugation at 20,000g, and the radioactivity of the soluble fraction was measured with a liquid scintillation counter. Purification of recombinant GST-PABPC1 was performed as described previously (Uchida et al. 2004)
Acknowledgments
We thank Dr. Kazuyoshi Yonezawa and Dr. Tadashi Yamamoto for plasmids. This work was supported in part by research grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and Grant-in-Aid for Research in Nagoya City University.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1597707
References
- Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T., Kobayashi T., Kajiho H., Hoshino S., Katada T., Kajiho H., Hoshino S., Katada T., Hoshino S., Katada T., Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchin C., Mauxion F., Sentis S., Seraphin B., Corbo L., Mauxion F., Sentis S., Seraphin B., Corbo L., Sentis S., Seraphin B., Corbo L., Seraphin B., Corbo L., Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Tarun S.Z., Boeck R., Sachs A.B., Tarun S.Z., Boeck R., Sachs A.B., Boeck R., Sachs A.B., Sachs A.B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Parker R., Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chen J., Chiang Y.C., Denis C.L., Chiang Y.C., Denis C.L., Denis C.L. CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G., Inge-Vechtomov S., Philippe M., Zhouravleva G., Philippe M., Zhouravleva G., Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion F., Seraphin B., Mauxion F., Seraphin B., Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Denis C.L., Chen J., Chen J. The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- Doma M.K., Parker R., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finoux A.L., Seraphin B., Seraphin B. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J. Biol. Chem. 2006;281:25940–25947. doi: 10.1074/jbc.M600132200. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., Guerrerio A.L., Parker R., Dietz H.C., Parker R., Dietz H.C., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Frolova L., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Drugeon G., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Kress M., Arman I., Haenni A.L., Celis J.E., Philippe M., Arman I., Haenni A.L., Celis J.E., Philippe M., Haenni A.L., Celis J.E., Philippe M., Celis J.E., Philippe M., Philippe M., et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Frolova L., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L., Zhouravleva G., Davydova E., Philippe M., Kisselev L., Davydova E., Philippe M., Kisselev L., Philippe M., Kisselev L., Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M., Hook B.A., Seay D.J., Wickens M., Seay D.J., Wickens M., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V., Zavialov A., Kisselev L., Ehrenberg M., Zavialov A., Kisselev L., Ehrenberg M., Kisselev L., Ehrenberg M., Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Miyazawa H., Enomoto T., Hanaoka F., Kikuchi Y., Kikuchi A., Ui M., Miyazawa H., Enomoto T., Hanaoka F., Kikuchi Y., Kikuchi A., Ui M., Enomoto T., Hanaoka F., Kikuchi Y., Kikuchi A., Ui M., Hanaoka F., Kikuchi Y., Kikuchi A., Ui M., Kikuchi Y., Kikuchi A., Ui M., Kikuchi A., Ui M., Ui M. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 1989;8:3807–3814. doi: 10.1002/j.1460-2075.1989.tb08558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S., Imai M., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T., Imai M., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T., Mizutani M., Kikuchi Y., Hanaoka F., Ui M., Katada T., Kikuchi Y., Hanaoka F., Ui M., Katada T., Hanaoka F., Ui M., Katada T., Ui M., Katada T., Katada T. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors. Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 1998;273:22254–22259. doi: 10.1074/jbc.273.35.22254. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai M., Kobayashi T., Uchida N., Katada T., Imai M., Kobayashi T., Uchida N., Katada T., Kobayashi T., Uchida N., Katada T., Uchida N., Katada T., Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T., Kikuchi Y., Hoshino S., Katada T., Hoshino S., Katada T., Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- Ikematsu N., Yoshida Y., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T., Yoshida Y., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T., Onda M., Hirai M., Fujimoto J., Yamamoto T., Hirai M., Fujimoto J., Yamamoto T., Fujimoto J., Yamamoto T., Yamamoto T. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Ohno M., Dreyfuss G., Ohno S., Dreyfuss G., Ohno S., Ohno S. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K.M., Salas-Marco J., Osherovich L.Z., Bedwell D.M., Salas-Marco J., Osherovich L.Z., Bedwell D.M., Osherovich L.Z., Bedwell D.M., Bedwell D.M. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Funakoshi Y., Hoshino S., Katada T., Funakoshi Y., Hoshino S., Katada T., Hoshino S., Katada T., Katada T. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 2004;279:45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- Kozlov G., Trempe J.F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K., Trempe J.F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K., Kahvejian A., Ekiel I., Gehring K., Ekiel I., Gehring K., Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N.S., Kozlov G., Chang T.C., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., Kozlov G., Chang T.C., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., Chang T.C., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., Siddiqui N., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., Volpon L., De Crescenzo G., Shyu A.B., Gehring K., De Crescenzo G., Shyu A.B., Gehring K., Shyu A.B., Gehring K., Gehring K. Comparative peptide binding studies of the PABC domains from the ubiquitin–protein isopeptide ligase HYD and poly(A)-binding protein. J. Biol. Chem. 2006;281:14376–14382. doi: 10.1074/jbc.M600307200. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Evans M.C., Agrin N.S., Smith M., Gongidi P., Jacobson A., Evans M.C., Agrin N.S., Smith M., Gongidi P., Jacobson A., Agrin N.S., Smith M., Gongidi P., Jacobson A., Smith M., Gongidi P., Jacobson A., Gongidi P., Jacobson A., Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes K.C., Wilusz C.J., Wilusz J., Wilusz C.J., Wilusz J., Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes & Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Okochi K., Suzuki T., Inoue J., Matsuda S., Yamamoto T., Suzuki T., Inoue J., Matsuda S., Yamamoto T., Inoue J., Matsuda S., Yamamoto T., Matsuda S., Yamamoto T., Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein. Genes Cells. 2005;10:151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Deardorff J.A., Deardorff J.A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Semotok J.L., Cooperstock R.L., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A., Cooperstock R.L., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A., Vari H.K., Lipshitz H.D., Smibert C.A., Lipshitz H.D., Smibert C.A., Smibert C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Siddiqui N., Mangus D.A., Chang T.C., Palermino J.M., Shyu A.B., Gehring K., Mangus D.A., Chang T.C., Palermino J.M., Shyu A.B., Gehring K., Chang T.C., Palermino J.M., Shyu A.B., Gehring K., Palermino J.M., Shyu A.B., Gehring K., Shyu A.B., Gehring K., Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Cox B.S., Ter-Avanesyan M.D., Tuite M.F., Ter-Avanesyan M.D., Tuite M.F., Tuite M.F. The products of the SUP45 and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Belasco J.G., Greenberg M.E., Belasco J.G., Greenberg M.E., Greenberg M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Thore S., Mauxion F., Seraphin B., Suck D., Mauxion F., Seraphin B., Suck D., Seraphin B., Suck D., Suck D. X-ray structure and activity of the yeast Pop2 protein: A nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R., Staples R.R., Chen J., Denis C.L., Parker R., Chen J., Denis C.L., Parker R., Denis C.L., Parker R., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R., Valencia-Sanchez M.A., Muhlrad D., Parker R., Muhlrad D., Parker R., Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Hoshino S., Imataka H., Sonenberg N., Katada T., Hoshino S., Imataka H., Sonenberg N., Katada T., Imataka H., Sonenberg N., Katada T., Sonenberg N., Katada T., Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Uchida N., Hoshino S., Katada T., Hoshino S., Katada T., Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Staples R.R., Baker R.E., Parker R., Staples R.R., Baker R.E., Parker R., Baker R.E., Parker R., Parker R. Function of the ski4p and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R., Frischmeyer P.A., Dietz H.C., Parker R., Dietz H.C., Parker R., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Viswanathan P., Ohn T., Chiang Y.C., Chen J., Denis C.L., Ohn T., Chiang Y.C., Chen J., Denis C.L., Chiang Y.C., Chen J., Denis C.L., Chen J., Denis C.L., Denis C.L. Mouse CAF1 can function as a processive deadenylase/3′–5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- Wang W., Czaplinski K., Rao Y., Peltz S.W., Czaplinski K., Rao Y., Peltz S.W., Rao Y., Peltz S.W., Peltz S.W. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Zhong Z., Chen C.Y., Shyu A.B., Chen C.Y., Shyu A.B., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M., Inge-Vechtomov S., Kisselev L., Philippe M., Kisselev L., Philippe M., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]