Abstract
The subgenomic mRNA of feline caliciviruses is bicistronic with the two cistrons overlapping by four nucleotides, .AUGA. The upstream cistron encodes a 75-kDa major capsid protein precursor (pre-VP1), and the downstream cistron a 10-kDa minor capsid protein. The kinetics of translation in reticulocyte lysates show that the downstream cistron is translated by a termination–reinitiation process, which is unusual in not requiring eIF4G or the eIF4F complex. Reinitiation requires the 3′-terminal 87 nucleotides (nt) of the pre-VP1 ORF, but no other viral sequences. The reinitiation site is selected by virtue of its proximity to this 87-nt element, and not its proximity to the pre-VP1 ORF stop codon, although this must be located not more than ∼30 nt downstream from the restart codon. This 87-nt element was shown to bind 40S ribosomal subunits and initiation factor eIF3, and addition of supplementary eIF3 enhanced reinitiation efficiency. Mutants defective in reinitiation showed reduced affinity for eIF3 or defective 40S subunit binding (or both). These results suggest a mechanism in which some of the eIF3/40S complexes formed during disassembly of post-termination ribosomes bind to this 87-nt element in a position appropriate for reinitiation following acquisition of an eIF2/GTP/Met-tRNAi ternary complex.
[Keywords: Initiation factor eIF3, mammalian mRNA translation, reinitiation, termination]
It is widely agreed that mammalian ribosomes generally do not, and cannot, translate the downstream cistron of a bicistronic mRNA (in sharp contrast to what happens in prokaryotic systems). There are two exceptions to this general rule. One is where the intercistronic spacer includes an IRES (internal ribosome entry site/segment) capable of recruiting ribosomes directly to the downstream cistron. The other is when the first ORF is short (less than ∼30 codons), and if eIF4F (or at least the central domain of eIF4G together with eIF4A) participated in initiation of translation of this short ORF, in which case some ribosomes that have completed translation of this short cistron may resume scanning in a 5′ → 3′ direction and reinitiate translation at a downstream AUG codon (Kozak 2001; Pöyry et al. 2004).
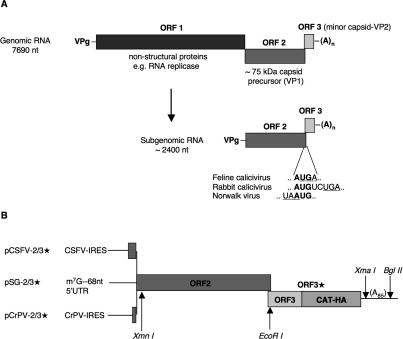
Nevertheless, there are a few cases of apparent reinitiation after a long ORF, which, if true, raises the important question: What is the special mechanism that promotes reinitiation in these rather exceptional cases, but not with most mRNAs? One such exception is the bicistronic subgenomic RNAs of the Caliciviridae family of positive-strand RNA viruses, including feline calicivirus (FCV), the species we chose for study. FCV virion RNA is ∼7700 nucleotides (nt) long and has three ORFs. It is thought that translation of the virion RNA leads to expression of only the 5′-proximal ORF1, encoding a polyprotein precursor of all the nonstructural proteins (Fig. 1A), which include an RNA replicase, a VPg, and a protease that processes both the ORF1 polyprotein product and the major capsid protein precursor (Wirblich et al. 1996; Clarke and Lambden 2000). The RNA replicase not only synthesizes more full-length viral RNA, but also a single species of ∼2400-nt subgenomic mRNA (Fig. 1A; Herbert et al. 1996), which is bicistronic, with an upstream ORF (identical to ORF2 of the virion RNA) encoding an ∼75-kDa precursor of the major capsid protein, VP1, and a downstream cistron (corresponding to ORF3 of full-length viral RNA) coding for an ∼10-kDa minor capsid protein (VP2) that is present in the virion at one to two molecules per particle (compared with 180 molecules of VP1), and is essential for infectivity (Sosnovtsev and Green 2000; Sosnovtsev et al. 2005).
Figure 1.
Diagrammatic map of FCV genomic (virion) and subgenomic RNAs, and the constructs used in this investigation. (A) The top part depicts the ORF structure of the virion (genomic) RNA, with the ORF structure of the single species of subgenomic RNA shown below. The ORF2/ORF3 overlap sequence of FCV subgenomic RNA is given, together with with the largest (RHDV) and smallest (Norwalk virus) overlaps found in other caliciviruses. (B) Structure of the three starting constructs used in this study. All three have the complete FCV subgenomic RNA ORF2 and ORF3 sequences with the latter fused in-frame to a CAT/triple-HA-tag cistron (lacking the CAT AUG initiation codon). This extended ORF3 is designated ORF3*. The three constructs differ in the sequences upstream of ORF2. pSG-2/3* has a 68-nt 5′-UTR described previously (Pöyry et al. 2004), and the 5′-proximal sequences of pCSFV-2/3* and pCrPV-2/3* are described in the text. The positions of restriction enzyme sites mentioned in the text are shown.
An unusual feature of both the full-length and subgenomic RNAs of caliciviruses is that they do not have a 5′-cap, but a covalently linked ∼15-kDa protein (VPg), which is encoded within ORF1 (Herbert et al. 1997). Nevertheless it is thought that initiation on these RNAs is by a ribosome scanning mechanism virtually identical to that operating with conventional capped mRNAs, although obviously there must be some difference in how the 5′-end is recognized by initiation factors (Goodfellow et al. 2005). There is no evidence that the VPg fulfills a role that is distinctly different from the usual 5′-cap, and, indeed, full-length capped transcripts are infectious (Sosnovtsev and Green 1995; Sosnovtsev et al. 2005).
The two ORFs of the single species of subgenomic mRNA overlap by four nucleotides in FCV (..AUGA..), and in other caliciviruses the overlap ranges from one nucleotide in Norwalk virus (..UAAUG..) to eight residues in rabbit hemorrhagic disease virus (Fig. 1A; Meyers 2003). Translation of this subgenomic RNA results in the synthesis of both proteins, suggestive of a termination–reinitiation event that is rather inefficient, with the VP2/pre-VP1 molar yield ratio given in one report as 0.1 (Herbert et al. 1996), but only just >0.05 in another study (Luttermann and Meyers 2007). The cis-acting RNA elements required for downstream cistron expression from both rabbit hemorrhagic disease virus (RHDV) and FCV subgenomic RNAs have been investigated in transfection assays (Meyers 2003, 2007; Luttermann and Meyers 2007). Expression of the downstream cistron was dependent on close proximity of the upstream cistron stop codon and downstream cistron (re)initiation codon. Second, downstream cistron expression required the 3′-terminal ∼90 nt of the upstream cistron, but no other viral sequences. Third, the putative reinitiation event required translation through this terminal segment of the upstream cistron but was not compromised if most of this segment was translated in the wrong reading frame, provided the frameshift was corrected just before the termination codon (Meyers 2003).
Because this previous work was based entirely on transfection assays, it was not possible to obtain direct evidence for a stop–restart mechanism, nor was it possible to investigate other mechanistic aspects, such as the requirements for translation initiation factors. The work reported here used in vitro translation assays to examine not only the requirement for cis-acting RNA motifs, with results consistent with the findings of Meyers (2003) and Luttermann and Meyers (2007), but also to examine these mechanistic aspects. Kinetic analysis shows that the expression of the downstream ORF does indeed involve a termination–reinitiation event, although one that is unusual in that it is independent of eIF4G or the eIF4F complex, but involves interaction of eIF3 and 40S ribosomal subunits with the 3′-terminal ∼90 nt of the upstream ORF. In view of the recently discovered function of eIF3 in releasing the post-termination ribosome from the mRNA as a eIF3/40S complex and a 60S ribosomal subunit (Pisarev et al. 2007), these results suggest a mechanism in which the binding of some post-termination eIF3/40S complexes to this short element of the FCV subgenomic mRNA captures and restrains them in a position suitable for reinitiation once they have acquired an eIF2/GTP/Met-tRNAi ternary complex. This requirement for a specific cis-acting RNA element with the property of binding eIF3 and 40S subunits can explain why termination–reinitiation events after translation of a long ORF do not generally occur with the overwhelming majority of mammalian mRNAs.
Results
Characterization of ORF3 expression in reticulocyte lysate
To facilitate comparison with the recent work of Luttermann and Meyers (2007), we used their nomenclature, designating the two cistrons of the FCV subgenomic mRNA as ORF2 and ORF3, but it should be noted that this is not the same as we used in a previous review article (Jackson et al. 2007), where our nomenclature conformed to the different scheme used by Meyers (2003) for RHDV subgenomic mRNA.
All starting constructs included FCV coding sequences from the start of ORF2 to the 3′-end of ORF3, modified by extending ORF3 with CAT coding sequences (lacking the initiation codon) followed by a triple HA-tag (Fig. 1B), to facilitate ORF3 product identification quantitation and resolution on SDS-PAGE. Rather than use the 17-nt natural FCV subgenomic mRNA 5′-untranslated region (UTR), which might be prone to leaky scanning due to its short length (Kozak 1991), a 68-nt 5′-UTR, described previously (Pöyry et al. 2004), was inserted ahead of ORF2. This construct is designated pSG-2/3* (with the 3* signifying that ORF3 is extended by CAT/triple-HA sequences).
We also replaced this 5′-UTR by nucleotides 1–423 of classical swine fever virus followed by an ACC linker, generating pCSFV-2/3*. This represents the entire CSFV 5′-UTR (including the IRES), plus the first 17 codons of viral coding sequence, which were included because they enhance internal initiation efficiency in some situations (Fletcher et al. 2002). In a third type of construct (pCrPV-2/3*), the 5′-UTR element of pSG-2/3* was replaced by the cricket paralysis virus intergenic IRES plus the first five codons of downstream viral coding sequence followed by an ACC linker (Pöyry et al. 2004). Linearization of the plasmids with either BglII or XmaI allowed the production of mRNAs with or without a poly(A) tail, but as there was no difference in the ORF3*/ORF2 product yield ratio (Supplementary Fig. S1), most experiments were done with poly(A)-RNA. All three types of transcript were translated under the same conditions, except for the concentration of added KCl, which was the standard 100 mM for capped transcripts of pSG-2/3*, but the optimum 120 mM KCl for the two uncapped IRES-containing RNAs.
The ORF3* (VP2-CAT-HA3) product, migrating at ∼43 kDa as expected, was identified by immunoprecipitation using anti-HA antibodies, and by its absence in assays of a transcript of pSG-2/3* that had been linearized with EcoRI at the very start of ORF3* (Supplementary Fig. S1). We also translated capped transcripts of pSG-2/3* that had been linearized with XmnI near the start of ORF2 (Fig. 1B), but no ORF3* product was detected (Supplementary Fig. S1), an important control showing that there is no cryptic T7 promoter within ORF2 generating a monocistronic mRNA that could give rise to ORF3* expression via 5′-end-dependent scanning. In addition, RNA (re)extracted from translation assays of the full-length transcript was analyzed by gel electrophoresis, but no cleaved RNAs of a size that could give rise to the ORF3* product by scanning were detected (Supplementary Fig. S2).
Time-course experiments confirm that initiation of translation of the downstream cistron is delayed until upstream cistron translation has been completed
In time-course experiments using an uncapped transcript of pCSFV-2/3*, the ∼75-kDa ORF2 product first appeared at 12 min, much as expected from our previous estimates of the rate of translation elongation in reticulocyte lysates (Jackson 1986), but the smaller ORF3* product did not appear until 21 min (Fig. 2A). This unexpectedly late appearance of the ORF3* product was not due to an abnormally slow rate of elongation through ORF3*, but to a delay in initiation of ORF3* translation, as shown by the fact that almost no ORF3* product was synthesized if the initiation inhibitor, edeine, was added at any time before ∼10 min (Fig. 2B). If edeine addition was delayed until after 12 min, the ORF3* product was detectable, with the ORF3*/ORF2 product yield ratio increasing the later the time of edeine addition (data not shown).
Figure 2.
Time course of translation of the surrogate FCV subgenomic RNA. (A) Uncapped CSFV-2/3* mRNA was translated under standard conditions. Samples were taken at the times indicated for separation by gel electrophoresis, and the resulting autoradiograph is shown. (B) Products of translation of uncapped CSFV-2/3* mRNA (at 80 μg/mL) for 60 min, with (+) or without (−) addition of edeine to 2 μM at 8 min. (C) Products of translation of SG-2/3* and CSFV-2/3* mRNAs in standard RRL (un) and eIF4G-depleted lysate (d).
Reinitiation occurs by a mechanism that is sensitive to inhibition by edeine but independent of eIF4G
The results of Figure 2B imply that the reinitiation mechanism leading to ORF3* translation is sensitive to inhibition by edeine. With mRNAs translated via the scanning mechanism, edeine does not inhibit the binding of an eIF2/GTP/Met-tRNAi ternary complex to the 40S ribosomal subunit, nor Met-tRNAi/40S complex scanning, but there is a complete failure of AUG codon recognition, so that scanning continues past all AUG codons, and, probably as a secondary consequence, there is no ribosomal subunit joining (Kozak and Shatkin 1978; Kozak 1979).
Although reinitiation appears to be via the standard mechanism by the criterion of sensitivity to edeine, it is decidedly nonstandard in being completely independent of eIF4G or the eIF4F complex. This was demonstrated by using the eIF4G-depleted reticulocyte lysate system described previously (Ali et al. 2001) and an uncapped transcript where upstream ORF2 expression is driven by the CSFV IRES, which does not require eIF4F or eIF4G in any form (Pestova et al. 1998). The synthesis of the ORF3* product was just as efficient in this system as in a standard reticulocyte lysate (Fig. 2C). Moreover, standard RRL assays of capped SG-2/3* mRNA that were supplemented at 6 min with recombinant dominant-negative R362 → Q mutant eIF4A (Pause et al. 1994; Pestova et al. 1998; Pöyry et al. 2004) showed that this independence of eIF4F (or eIF4G/4A) was not a peculiarity of using an RNA with the CSFV IRES. As expected, the dominant-negative eIF4A severely curtailed further initiation of ORF2 translation, but the ORF3*/ORF2 product yield ratio was the same as in an unsupplemented control (data not shown), thus demonstrating that those ribosomes that initiated ORF2 translation in the first 6 min reinitiated at the ORF3* restart codon with normal probability, despite the dominant-negative eIF4A. With the exception of initiation promoted by the pestivirus, hepatitis C virus, and CrPV IRESs (Pestova et al. 1998; Wilson et al. 2000; Pestova and Hellen 2003), and scanning-dependent mRNAs with an absolutely unstructured 5′-UTR (Pestova and Kolupaeva 2002), there are no precedents for initiation without the involvement of eIF4G in some form, either as intact eIF4G in the eIF4F complex, or, at a minimum, the central one-third domain of eIF4G, which retains the potential to interact with eIF3 and eIF4A (for review, see Jackson 2005).
The data of Figure 2C clearly show that reinitiation efficiency in the standard RRL system is slightly lower in the case of the mRNA with the CSFV-IRES than capped SG-2/3* mRNA. Over several different experiments, the ORF3*/ORF2 molar yield ratio was in the range 0.10–0.15 for capped SG-2/3* mRNAs, and 0.08–0.12 for CSFV-2/3* mRNA. Our results suggest that two factors contribute to this lower efficiency. One is the higher concentration of added KCl in the assays of the IRES-containing mRNA. The other is the fact that the CSFV IRES binds a translation initiation factor (eIF3), which, as we show subsequently, is directly involved in the reinitiation mechanism.
Deletion mapping of the cis-acting RNA element required for the termination–reinitiation event
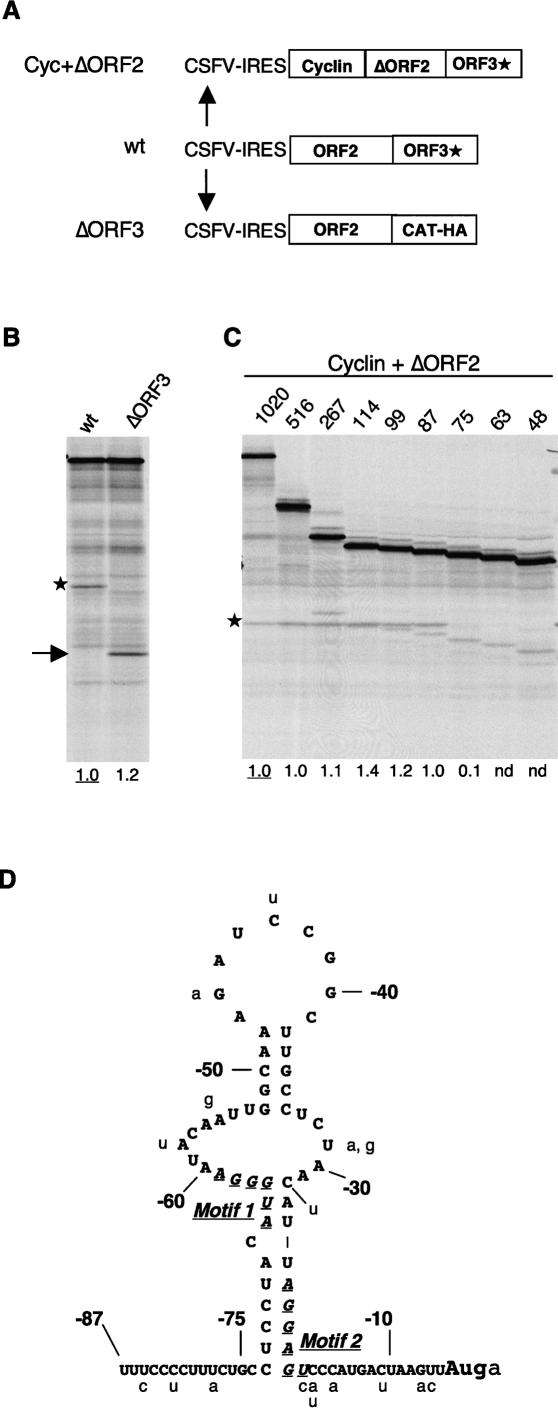
To investigate the requirement for viral sequences downstream from the reinitiation site, the whole of the viral ORF3 coding sequences (apart from the first 7 nt) were deleted and replaced by an AUGAGCUC linker (Fig. 3A). The lack of any reduction in downstream CAT-HA3 cistron expression (Fig. 3B) shows that viral ORF3 sequences are not required for the reinitiation event.
Figure 3.
Mapping the viral sequences required for downstream ORF expression. (A) Schematic diagram (not to scale) of the deletions introduced to map the requirement for viral sequences. (B) Translation of the ΔORF3 mRNA shows that ORF3 coding sequences are not required for downstream cistron expression. The full-length ORF3* product is indicated by an asterisk, and the CAT-HA product is indicated by an arrow. (C) Deletion mapping of the ORF2 sequences required for ORF3* expression. The deletions are designated according to the number of 3′-terminal residues of ORF2 retained. The ORF3* product is indicated by an asterisk. Numbers below each lane show the efficiency of reinitiation (the ORF3*/ORF2 product yield ratio) expressed relative to the control mRNA value set at 1.0. (nd) Not detectable above background (i.e., effectively zero). The anomalous high value for the construct with 114 nt retained is due to the comigration of the ORF3* product with an incomplete translation product of ORF2. (D) Sequence and tentative secondary structure prediction of the 3′-terminal 87 nt of FCV strain F9 ORF2, with the variations found in other strains shown in lowercase. The core segments of Motif 1 and Motif 2 identified by Luttermann and Meyers (2007) as especially critical for downstream cistron translation are highlighted by italics and underlining.
The requirement for upstream viral sequences was investigated by making a nested set of 5′-deletions of ORF2, and fusing the residual ORF2 segment in-frame to the Xenopus laevis cyclin B2 complete ORF (lacking a termination codon), in the background of pCSFV-2/3* (Fig. 3A). Retention of the 3′-terminal 87 nt (i.e., the terminal 28 sense codons plus the stop codon) of ORF2 was found to give the same ORF3*/ORF2 product yield ratio as the full-length complete ORF2; if only the 3′-terminal 75 nt of ORF2 was retained, the yield ratio was consistently in the range 10%–15% of this control value, and the ORF3* product was undetectable if just the terminal 63 nt was retained (Fig. 3C).
For comparison, the transfected cell assays of Luttermann and Meyers (2007) gave 75%–80% of maximum reinitiation efficiency when the terminal 84 or 72 nt was retained, 30% with the terminal 69 nt, and essentially zero ORF3 expression with just the terminal 66 nt of FCV ORF2. Thus the main discrepancy with our results concerns their 72- and our 75-nt truncations, suggesting that the minimum length of required 3′-terminal ORF2 sequences may be greater for in vitro translation assays than transfected cells. This difference is certainly due to the higher protein concentration (notably the eIF3 concentration) in intact cells, because, as shown later, supplementation of our in vitro translation assays with extra eIF3 stimulated reinitiation with the 75-nt truncation mutant by six- to sevenfold, thus completely eliminating the difference between in vitro and transfected cell assays,
The sequence of the 3′-terminal 87 nt of FCV (F9 strain) ORF2 is shown in Figure 3D, together with a tentative secondary structure prediction based on a combination of Mfold predictions and direct structure probing of a 5′-end-labeled fragment representing the terminal 114 nt of ORF2 (data not shown). Phylogenetic comparison of different FCV strains did not contribute much additional information except to show that almost all variations were in segments predicted to be unpaired (Fig. 3D), while there is too much divergence between different species of the Caliciviridae for cross-species comparisons to be useful. We did not carry out more detailed mapping of this essential 87-nt segment, as in the meantime this was done by Luttermann and Meyers (2007) for the FCV system, with a useful comparative study of the rabbit calicivirus (RHDV) by Meyers (2007). In both cases, two short segments, designated Motif 1 and Motif 2 (see Fig. 3D), were found to be particularly important for downstream cistron expression. The spacing, but not the sequence, between Motif 2 and the restart site was critical for reinitiation, but shortening the distance between Motifs 1 and 2 had only a small effect. Motif 1 has an essential A/GUGGGA core sequence (Fig. 3D) that is conserved in other caliciviruses. In addition, nonconserved flanking sequences up to six residues on either side of this core seem to play a stimulatory rather than an essential role. In both cases, Motif 2 has a core 6 nt in length that, apart from being purine-rich, is not conserved between FCV and RHDV. Consequently, the RHDV Motif 2 could only be identified by experimental mutagenesis, and not by simple sequence comparison with the corresponding region of FCV RNA (Meyers 2007). The lack of conservation in Motif 2 and in the flanking sequences of Motif 1 suggests that these elements may required more for secondary structure reasons than as primary sequence signals.
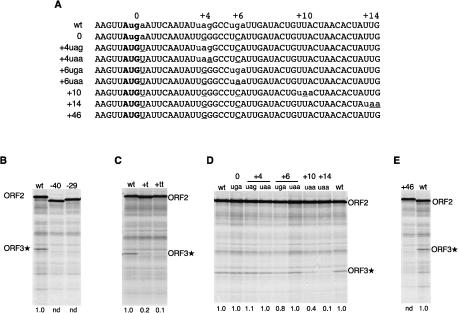
The importance of the proximity of the ORF2 stop codon to the ORF3 initiation codon
The position of the ORF2 termination codon was altered, while maintaining the ORF3* initiation codon in its original position. Introduction of a UAA codon 29 or 40 codons upstream of the endogenous ORF2 stop codon completely abrogated ORF3* expression (Fig. 4B). Because the sequence of at least part of the 3′-terminal 87 nt of ORF2 is important for reinitiation (as we show below), but the reading frame is not critical (Meyers 2003), we used frameshift rather than point mutations to engineer premature termination within this segment, by inserting one or two T residues immediately upstream of the −87 position in pCSFV-2/3*. The single insertion generated an in-frame UAA 28 nt upstream of the endogenous stop codon residues −31 to −29 in Figure 3D, which reduced reinitiation by 80%; and the double insertion created an in-frame UGA at −14/12 (Fig. 3D), which caused a 90% decrease in ORF3* expression (Fig. 4C).
Figure 4.
The effect on ORF3* expression of displacing the ORF2 termination codon. (A) Sequences of the ORF2/ORF3* overlap region and the start of ORF3* in the transcripts of the various mutants of pCSFV-2/3* tested. The ORF3* initiation codon is in bold, termination codons are in lowercase, and the introduced mutations are underlined. (B) Translation products of CSFV-2/3* mRNA with UAA stop codons introduced in the ORF2 frame 29 or 40 codons upstream of the wild-type ORF2 stop codon. (C) Products of translation of transcripts of pCSFV-2/3* with one (t) or two (tt) T residues inserted into ORF2 between residues −87 and −88 in the numbering system of Figure 3D. (D) Translation products of the CSFV-2/3* mutants with the ORF2 stop codon displaced up to 14 codons further downstream, as shown in A. (E) Translation products of CSFV-2/3* mRNA lacking the wild-type ORF2 termination codon and the in-frame stop codons at +4 and +6 (shown in A), resulting in an extension of ORF2 by 46 codons. The numbers below each lane show the ORF3*/ORF2 product yield ratio (reinitiation efficiency) obtained with each mutant relative to the wild-type mRNA value set at 1.0. (nd) Not detectable above background (i.e., effectively zero).
To study the effect of extending ORF2 by moving the termination codon further downstream, we initially took advantage of the existence of in-frame stop codons located four and six codons downstream from the natural termination codon and made each of these the sole termination codon in this region (Fig. 4A). The efficiency of ORF3* expression was largely unaffected by having the sole stop codon at the +4 (UAG) or +6 (UGA) position, and having a UAA stop codon at these positions also had no effect (Fig. 4D).
No ORF3* expression whatsoever occurred when all three of these stop codons were mutated, which resulted in an extension of ORF2 by 46 codons (Fig. 4E). When an in-frame UAA codon was introduced at the +10 or +14 position (in the background of a construct in which the endogenous stop codon and the +4 and +6 termination codons had all been mutated to sense codons), the ORF3*/ORF2 product yield ratio was consistently reduced by 50%–60% with the 10-codon extension, and by 90% with the extra 14 codons (Fig. 4D). Thus reinitiation occurs only if ORF2 translation terminates within a narrow window extending ∼10 codons downstream from the wild-type stop codon position. Notably, the expression of ORF3* when ORF2 was extended by four, six, or 10 codons was as completely resistant to inhibition by R362 → Q dominant-negative eIF4A as when ORF2 terminated at the endogenous stop codon (data not shown).
What determines the selection of the reinitiation codon?
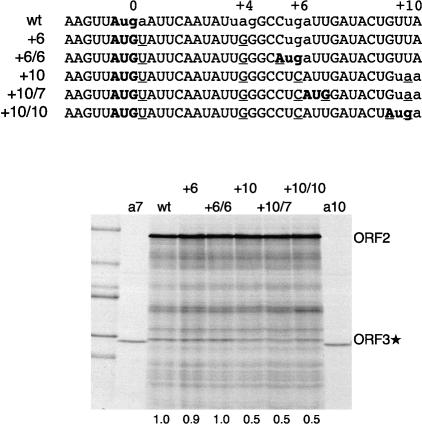
In order to test whether the termination–reinitiation mechanism selects the ORF3* initiation codon by virtue of its proximity to the ORF2 termination codon, we took constructs in which the ORF2 termination codon had been displaced further downstream and created a new AUG codon in the ORF3* reading frame, as an alternative and competing (re)initiation site in the extended region of overlap. Three such mutants were generated (in the background of pCSFV-2/3*) with the sequences shown in Figure 5, some of which recapitulated the ..-AUGA.. overlap found in the wild-type stop–restart site. When these mRNAs were translated in vitro, no products initiated at the newly created AUG codons were detected, but the usual ORF3* product initiated at the wild-type start codon was still synthesized in undiminished yield (Fig. 5), consistent with the conclusion that there was no competing initiation at the new AUGs. We conclude that the reinitiation site is selected by virtue of its proximity to the critical 87-nt 3′-terminal element of ORF2, and not its proximity to the ORF2 stop codon, even though this stop codon must be located within a quite narrow window relative to the restart site.
Figure 5.
The restart codon is selected by virtue of its proximity to the ORF2 87-nt element and not by its proximity to the ORF2 stop codon. The top part shows the sequences of the pCSFV-2/3* mutants tested, with initiation codons in the ORF3* frame shown in bold, termination codons in the ORF2 frame shown in lowercase, and the introduced mutations underlined. The bottom panel shows the products of translation of these mRNAs (at 80 μg/mL). The outside lanes designated “a7” and “a10” show products of translation of capped monocistronic mRNAs with a start codon in either the +7 or +10 position and 5′-UTRs of 21 or 30 nt in length, respectively, and serve as markers for any initiation at these two sites that might have occurred (but did not actually occur) in translation assays of the CSFV-2/3* mRNAs. Numbers below each lane show the relative reinitiation efficiency determined and defined as in Figure 4.
Thus it seems that the wild-type position of the ORF3 initiation codon is somehow a “privileged site” for reinitiation. A further indication of this was seen when the ORF2 stop codon was mutated to UAA, which consequently resulted in the reinitiation codon becoming AUA. Despite this change to a non-AUG codon that seldom functions as an initiation codon in other mRNAs, the yield of ORF3* product was reduced by only ∼65% (data not shown). Similarly, Luttermann and Meyers (2007) found that expression of FCV ORF3 was reduced by only 50%–60% when the AUG restart codon was mutated to AUA, CUG, or GUG; even CUA, which has never been found to act as an initiation codon in other mRNAs, allowed ORF3 expression at ∼30% of the efficiency seen with an AUG. With the exception of initiation promoted by the pestivirus, hepatitis C virus, and CrPV IRESs (Reynolds et al. 1995; Wilson et al. 2000), such efficient initiation at non-AUG codons is unprecedented, because eIF1 usually prevents recognition of such codons as initiation sites (Lomakin et al. 2006). This raises the interesting question of whether the reinitiation event occurs without the involvement of eIF1.
The terminal 87 nt of ORF2 binds eIF3 and 40S ribosomal subunits
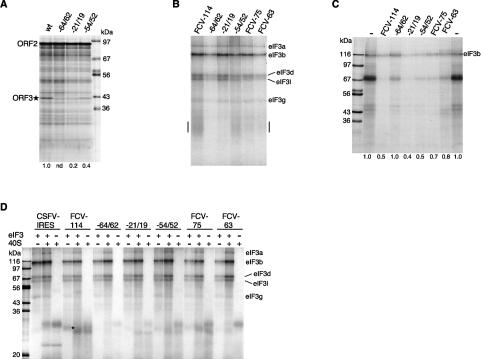
In order to understand how the ORF2 3′-terminal element promotes reinitiation, it would be useful to know what binds to it, particularly in a dynamic situation where ongoing translation termination is occurring at the end of this element. However, practical considerations necessitated studying protein–RNA interactions under static conditions, effectively treating the element as if it were an IRES. Although the translation data show that it has no detectable IRES activity, it seemed possible that the critical proteins might nevertheless bind, albeit weakly, under static conditions. Because only weak interactions were expected, we preferred UV-cross-linking over band-shift assays (for reasons of irreversibility), and included Escherichia coli rRNA in all assays to suppress nonspecific binding to the probe. The pattern of cross-linked proteins obtained with the terminal 114 nt of FCV ORF2 using a HeLa cell cytoplasmic extract was remarkably similar to that seen with a CSFV IRES probe (Supplementary Fig. S3A,B), and virtually identical results were seen using RRL except that the signal intensity of all bands was lower. Since the CSFV IRES is known to bind eIF3 (Pestova et al. 1998; Sizova et al. 1998), these UV-cross-linking reactions were subjected to immunoprecipitation with anti-eIF3, and in addition, cross-linking assays were done with purified eIF3. Both approaches confirmed that eIF3a, eIF3b, eIF3d, eIF3l, and eIF3g were cross-linked to the FCV element (Supplementary Fig. S3C,D,E), showing that this element binds not only purified eIF3 but also the eIF3 present in HeLa cell cytoplasmic extracts. (The HeLa cell extracts clearly have several other proteins that bind to the FCV ORF2 terminal segment, but these show no obvious correspondence to known initiation factors.)
We next examined several mutants of the ORF2 3′-terminal element for correlations between reinitiation efficiency and the binding of eIF3 to this segment. The mutants included not only the truncations of the cis-acting element to 75 or 63 nt examined in Figure 3C, but also the following point mutations in the pCSFV-2/3* background: GGG−64/62 → CCC in Motif 1 of Luttermann and Meyers (2007), AGU−21/19 → UUA in Motif 2, and UUG−54/52 → AAU (see Fig. 3D). The −54/52 mutant consistently retained 35%–40% of the control reinitiation activity, and the −21/19 mutant 15%–20%, but the GGG−64/62 → CCC mutant was essentially inactive (Figs. 6A, 7C). Taken together with the results of Figure 3C, the following hierarchy of reinitiation efficiency was seen: “wild type” (at least the terminal 87 nt of ORF2 retained) ≫ −54/52 mutant ≫ −21/19 mutant > truncation to 75 nt ≫ 63-nt truncation ≈ −64/62 mutant, which were both judged to be completely inactive.
Figure 6.
Correlations between reinitiation efficiency and the eIF3 or 40S binding of various mutant derivatives of the 3′-terminal ORF2 element. (A) Translation assays of uncapped transcripts of pCSFV-2/3* wild-type control, and (in the same background) GGG−64/62 → CCC, AGU−21/19 → UUA, and UUG−54/52 → AAU mutants. Numbers below each lane show the relative reinitiation efficiency determined and defined as in Figure 4. (B) UV-cross-linking assays of purified eIF3 (200 nM) with the following labeled probes: the 3′-terminal 114 nt of ORF2 wild type and the three point mutants described above, and the 75-nt and 63-nt truncations. The bar in the margins highlights the smeared signal at ∼29 kDa discussed in the text. (C) UV-cross-linking competition assays with HeLa cell extract, the wild-type 114-nt 3′-terminal ORF2 element as labeled probe, and a 200-fold molar excess of the same set of point mutants and truncation mutants as unlabeled competitors. Numbers below each lane show the intensity of the eIF3b signal relative to the average of the two outside lanes with no competitor (−) set at 1.0. (D) UV-cross-linking assays with 160 nM eIF3, or 80 nM purified 40S subunits (or both together), as indicated, and the same labeled probes as in B. In order to reveal the weak 40S subunit cross-linking signals, the autoradiogram was deliberately overexposed, which resulted in some loss of differential eIF3 signal intensity between the various mutants, due to film saturation. The cross-linking signal attributed to eIF3k in the assay of FCV-114 and eIF3 alone is highlighted with an asterisk to the right of the relevant lane.
Figure 7.
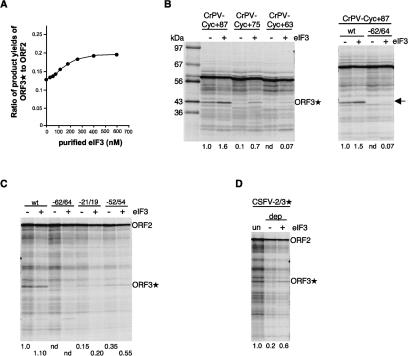
ORF3* expression is stimulated by supplementation of translation assays with eIF3. (A) CrPV-2/3* mRNA (100 μg/mL) was translated in the presence of varying concentrations of supplementary eIF3. The yields of the ORF3* and ORF2 products were determined by densitometry of the autoradiographs, and the molar yield ratio is plotted against the concentration of added eIF3. (B) Selected mRNAs from the deletion series shown in Figure 3C (but in the background of the construct with the CrPV IRES) and the −64/62 point mutation in the same background were translated (at 80 μg/mL) in the presence or absence of 250 nM supplementary eIF3. (C) Uncapped transcripts of full-length pCSFV-2/3*, or (in the same background) the −64/62, −54/52, and −21/19 point mutants were translated in the absence or presence of 250 nM supplementary eIF3. (D) Translation of CSFV-2/3* mRNA (80 μg/mL) in eIF3-depleted RRL (dep), with or without supplementation with 150 nM eIF3. Numbers below each lane show the relative ORF3*/ORF2 product yield ratio (i.e., reinitiation efficiency) determined and defined as in Figure 4.
These mutant sequences were then used as probes in UV-cross-linking assays with purified eIF3 (Fig. 6B), and as unlabeled competitors in cross-linking competition assays with HeLa cell lysate, the 114-nt terminal fragment of ORF2 as probe, and a single concentration of each mutant RNA element as competitor, monitoring the labeling of eIF3b (Fig. 6C). Taking all the results of these different assays into consideration, plus data (data not shown) from similar competition assays using purified eIF3, the consensus hierarchy of eIF3 binding to the mutant sequences was −21/19 mutant > wild-type control > −54/52 mutant > 75-nt truncation > 63-nt truncation > −64/62 mutant. In general, this rank order parallels the relative efficiency of reinitiation, but with the obvious exception of the −21/19 mutant, which binds eIF3 even more effectively than the control, yet gives only 15%–20% as much reinitiation (Figs. 6A, 7C). In other cross-linking assays with purified eIF3 and the wild-type control probe, we noted a band at ∼29 kDa (Fig. 6D), which was apparent as a smear in Figure 6B and which corresponds in position to eIF3k. The relative signal intensity of this band (wild-type ≫ −54/52 mutant > 75-nt truncation ≫ −21/19 mutant ≈ 63-nt truncation > −64/62 mutant, which gave essentially zero signal) correlated particularly well with reinitiation efficiency, with the slight reservation that the two truncation mutants gave somewhat stronger signals than would be expected.
We next attempted UV-cross-linking assays with purified 40S ribosomal subunits in the presence or absence of eIF3 (although the latter situation seldom, if ever, exists under physiological conditions). The presence of 40S subunits invariably increased the eIF3a, eIF3b, and eIF3d polypeptide cross-linking signals, and to a lesser extent eIF3l (Fig. 6D). Because this increase was seen with all mutant RNA elements, even those that showed virtually no 40S/mRNA cross-linking (e.g., GGG−64/62 → CCC), the signal enhancement seems more likely to be due to conformational changes induced in eIF3 when it interacts with 40S subunits than to stabilization of the eIF3/mRNA interactions by 40S/mRNA interactions. In contrast, the presence of eIF3 had little effect on the 40S subunit cross-linking signals obtained with the wild-type probe but decreased the intensity of labeling of many 40S-dependent bands in the case of the mutants (Fig. 6D), particularly those mutants that showed no detectable reinitiation (the 63-nt truncation and the −64/62 point mutant). We conclude that the reinitiation efficiency of all mutants, apart from the −21/19 point mutant, parallels the binding to the 3′-terminal segment of ORF2 of both eIF3 and 40S subunits (Fig. 6B–D); i.e., both components of the eIF3/40S complex that is the product of disassembly of the post-termination 80S ribosome (Pisarev et al. 2007). As for the −21/19 mutant, this appears to retain high affinity for eIF3 (Fig. 6B,C), although with very poor cross-linking of the putative eIF3k (Fig. 6D), but is defective for the binding of 40S subunits as judged by the fact that it does not give the same 40S-dependent labeled bands as the wild-type probe (Fig. 6D). This aberrant binding of 40S subunits provides a plausible explanation for its severely reduced reinitiation efficiency.
Functional evidence that eIF3 is involved in the reinitiation mechanism via binding to the 3′-terminal segment of ORF2
Given the reservations that the binding assays were unavoidably done under static conditions, it was considered essential to use functional tests to confirm the important role of eIF3 in reinitiation, and so we investigated the effect of manipulating the eIF3 concentration in translation assays. With the pCrPV-2/3* surrogate subgenomic mRNA, supplementary eIF3 stimulated ORF3* expression (with a negligible effect on ORF2 expression), resulting in a concentration-dependent increase in the ORF3*/ORF2 product yield ratio that reached a plateau value of 1.5-fold that of the control (unsupplemented) system (Fig. 7A). A similar concentration-dependent increase in ORF3*/ORF2 product yield ratio reaching a comparable plateau level although at a higher eIF3 concentration was seen with the other two surrogate subgenomic mRNAs (data not shown), but with the complication that especially with capped SG-2/3* mRNA, and to a lesser extent with uncapped CSFV-2/3* mRNA, supplementary eIF3 above ∼100 nM showed increasing inhibition of upstream ORF2 expression, for unknown reasons.
If it is the binding of eIF3 to the ∼90-nt 3′-terminal segment of ORF2 that is the critical eIF3 interaction required for reinitiation, a very strong prediction is that supplemental eIF3 should promote a stimulation proportionately >1.5-fold in the case of those mutants that show a moderately reduced binding of eIF3 (and reduced, but still detectable reinitiation). This was tested using mRNAs with an upstream cistron consisting of the cyclin B2 ORF fused to the 3′-terminal 87, 75, or 63 nt of ORF2 (Fig. 7B). With the terminal 87 nt retained, which is sufficient to promote full reinitiation efficiency (Fig. 3C), supplementary eIF3 stimulated ORF3* expression 1.5- to 1.6-fold (Fig. 7B), much as in Figure 7A. In contrast, with the deletion mutant in which only the terminal 75 nt of ORF2 was retained (resulting in an ∼10-fold reduction in reinitiation efficiency), supplementary eIF3 consistently elicited a much higher six- to sevenfold stimulation of the ORF3*/ORF2 product yield ratio (Fig. 7B), as is strongly predicted if reinitiation is dependent on eIF3 binding to this element.
Supplementary eIF3 also promoted a visible but extremely weak ORF3* band with the 63-nt truncation mutant, and the GGG−64/62 → CCC mutant (Fig. 7B), neither of which gave a detectable ORF3* product in the unsupplemented system. The effect of supplementary eIF3 on the two other point mutants was tested in the background of pCSVF-2/3* (Fig. 7C), where, as mentioned above, it causes some reduction in upstream ORF2 expression, and thus the results are not directly comparable with those of Figure 7B. The greatest stimulation in absolute terms was seen with the −54/52 mutant, which showed slightly lower eIF3 binding than the wild-type control in the majority of binding assays, as well as reduced binding of 40S subunits (Fig. 6D). With the −21/19 point mutant, which shows almost as low reinitiation efficiency as the 75-nt truncation, the proportional stimulation by supplementary eIF3 was greater than the wild type but was nevertheless rather modest, presumably because its main defect is in 40S subunit binding (Fig. 6D) rather than eIF3 binding, which is actually rather better than the wild type (Fig. 6B,C). Taken together, the results of Figure 6, B and C, argue strongly that binding of eIF3 to the terminal segment of ORF2 is a prerequisite for reinitiation and a major determinant of its efficiency, but is not sufficient in the absence of an appropriate interaction of the 40S subunits with this element.
We also attempted to deplete the system of endogenous eIF3 using oligo(dT) magnetic beads and a poly(A)-tailed derivative of the CSFV IRES with a CCA331–333 → GGU mutation in pseudoknot stem 1b that has been shown to abrogate translation and ribosome binding without perturbing eIF3 binding (Supplementary Fig. S4; Kolupaeva et al. 2000). This is similar to the approach that achieved ∼95% depletion of eIF4G from RRL in a single cycle (Ali et al. 2001). However, Western blotting showed that even after four cycles of exposure to the mutant CSFV IRES affinity matrix, only about one-third of the eIF3 had been removed (data not shown), suggesting that a large fraction of the endogenous eIF3 may be unavailable for binding to the affinity matrix; after four cycles, the translation efficiency of the lysate was distinctly compromised, presumably because other important factors had been partially removed. Nevertheless, this depletion reduced the ORF3*/ORF2 product yield ratio (the index of reinitiation efficiency) by fivefold, and supplementation of the depleted system with eIF3 resulted in (incomplete) rescue, raising the product yield ratio threefold (Fig. 7D). This confirms the critical role of eIF3 in the reinitiation mechanism and shows that the reduction in reinitiation resulting from the affinity depletion procedure is primarily due to depletion of eIF3 rather than some other proteins that happen to bind to the CSFV IRES.
Discussion
This investigation was driven by the wish to address the longstanding question: Why are termination–reinitiation events following translation of a long ORF so extremely rare in eukaryotic mRNA translation when they are commonplace with prokaryotic polycistronic mRNAs? To this end, we studied one of the few eukaryotic bicistronic mRNAs, calicivirus subgenomic mRNA, where downstream cistron translation appears to be dependent on such an obligatory termination–reinitiation event. Other bicistronic mRNAs possibly translated by a similar mechanism include the M2 mRNA of pneumoviruses such as respiratory syncytial virus (RSV), where the two ORFs overlap by 32 nt (Ahmadian et al. 2000; Gould and Easton 2005); the M RNA (segment 7) of the influenza B viruses, where the overlap is ..UAAUG.. (Horvath et al. 1990); and non-long-terminal repeat retrotransposons, the majority of which have either ..UAAUG.. or ..AUGA.. overlaps (Kojima et al. 2005). In all three cases, downstream cistron expression was absolutely dependent on translation of the upstream ORF and required close proximity of the termination and (re)start codons. Although the question of whether the putative reinitiation event required a cis-acting RNA element was not examined in the influenza B virus case, with RSV M2 mRNA, the 3′-terminal 150- to 200-nt segment of the upstream ORF was necessary, and in the case of the SART1 retrotransposon, it was suggested that a secondary structure element downstream from the restart site is required.
All our results are indicative of a termination–reinitiation mechanism for expression of FCV subgenomic mRNA ORF3. Taken together, the presence of 38 AUG triplets within the upstream ORF2, the time course of ORF3* expression (Fig. 2), plus the observations that ORF3* translation is dependent on the position of the ORF2 stop codon, and is sensitive to edeine but insensitive to dominant-negative mutant eIF4A when these are added before 10 min, eliminate most of the alternative explanations for ORF3* expression; e.g., leaky scanning, ribosome shunting, internal initiation via an IRES, or a frameshift leading to synthesis of an ORF2/ORF3* fusion protein that is subsequently cleaved.
This leaves only the rather baroque alternative that translation through the terminal ∼90 nt of ORF2 might remodel the RNA structure to generate a functional IRES. However, because ORF3* expression is abrogated if the ORF2 stop codon is moved more than ∼15 codons further downstream, it would be necessary to concede that the structure must snap back to the inactive conformation once the elongating ribosome has cleared this 90-nt segment, in which case would it not also snap back when a ribosome that has just terminated translation at the wild-type stop codon dissociates from the mRNA? Thus generation of a functional IRES by translation-dependent remodeling would only be a tenable hypothesis if the ribosome, or at least its small subunit, remained mRNA-associated following termination (in order to maintain the remodeled state), and this type of scenario is not an IRES within the usual meaning of that term.
In prokaryotic systems, this type of translational coupling involving an obligatory termination–reinitiation event is seen when the downstream cistron B initiation site has a vestigial Shine-Dalgarno (SD) motif that is too weak to recruit ribosomes from the free pool. Consequently, expression of cistron B is absolutely dependent on translation of the upstream cistron A and also requires close proximity of the cistron A stop codon and the cistron B (re)initiation site. As with the FCV system, the efficiency of reinitiation is low and is further reduced if the distance between the stop codon and the restart site is increased. Examples include reinitiation at sites within the lac i ORF and the T4 rII cistron B when there is a nearby in-frame nonsense mutation, and the translation of the bacteriophage f1 gene VII cistron, which is dependent on translation of the upstream gene V cistron (Sarabhai and Brenner 1967; Steege 1977; Ivey-Hoyle and Steege 1992). The data suggest that following termination at the cistron A stop codon, the ribosome, or more likely just its small subunit, undergoes bidirectional diffusion for a very limited period of time (and therefore limited in distance) before dissociating from the mRNA, unless it encounters a potential reinitiation site in this brief time window (Adhin and van Duin 1990). There is a strong preference for reinitiation at the nearest acceptable SD-initiation codon tandem (albeit one with a weak SD motif), even to the extent of preferring a nearby UUG start codon over a more distant AUG.
Kinetic analysis of the disassembly of the post-termination prokaryotic ribosome suggests that any such diffusion must be of very short duration indeed (Peske et al. 2005). The first step is the release of the large ribosomal subunit by the combined action of ribosome release factor (RRF), EFG, and GTP, leaving the small subunit (with an associated deacylated tRNA) still bound to the mRNA. Next, the binding of IF3 to the 30S subunit serves not only to prevent any rejoining of the large subunit but also promotes ejection of the tRNA, whereupon dissociation of the 30S subunit from the mRNA then follows almost immediately, leaving only the very briefest time for any bidirectional diffusion.
If the disassembly of the post-termination mammalian ribosome also leads to very limited bidirectional diffusion of the 40S subunit, the reason that termination–reinitiation events occur so rarely is presumably because there is no equivalent of the SD interaction to stabilize the association of this 40S subunit with the mRNA, and to restrain it in a position suitable for reinitiation once it had acquired an eIF2/GTP/Met-tRNAi ternary complex. A special type of reinitiation is seen if the upstream ORF is short (less than ∼30 codons) and translated rapidly without any holdups, and if, in addition, the eIF4F complex, or at least the central domain of eIF4G in association with eIF4A, participated in the primary initiation event at the upstream ORF initiation codon (Kozak 2001; Pöyry et al. 2004). Provided all these preconditions are met, some of the ribosomes that translated the upstream short ORF resume scanning and can reinitiate at a downstream AUG. The very nature of these preconditions suggests that the contacts between eIF4G and the ribosome, which is an indirect interaction bridged by eIF3 (Hinnebusch 2006), are not disrupted concomitantly with initiation at the short ORF AUG, but decay stochastically over a period of a few seconds following this initiation event; and that if this tripartite eIF4G/eIF3/ribosome interaction is still in place by the time translation of the short ORF is completed, it serves not only to retain the post-termination 40S subunit on the mRNA but also to promote (re)scanning. However, because this mechanism of retaining the post-termination 40S subunit on the mRNA is necessarily limited to very short ORFs, it cannot be operative in the case of FCV subgenomic mRNA. The radical difference between the two types of reinitiation is well illustrated by the fact that using a CSFV IRES to drive initiation of upstream ORF translation has no negative effect on reinitiation in the FCV system (Fig. 2C), but is nonpermissive for reinitiation downstream from a short ORF (Pöyry et al. 2004).
Given that reinitiation after translation of a long ORF is such a rare event in eukaryotes, it is intuitive that in the few exceptions there must be some cis-acting mRNA element that serves to retain and restrain some of the post-termination ribosomes (or more likely, just their 40S subunits) on the mRNA in the appropriate location relative to the reinitiation site. Our results confirm that the terminal ∼90 nt of ORF2 is essential for reinitiation to occur and that translation must proceed through this element and terminate not >10–15 codons downstream from the normal (wild-type) stop codon position. However, the reinitiation site is actually specified by its position relative to the 3′-terminal element of ORF2, not its position relative to the ORF2 stop codon. This 3′-terminal element of ORF2 has the potential to bind both 40S ribosomal subunits and eIF3 (Fig. 6). Because point mutations and truncations of this element that have a negative effect on reinitiation were either shown to be defective in eIF3 binding or showed aberrant interactions with 40S subunits (or to have both defects), it is likely that reinitiation depends on an appropriate interaction of post-termination eIF3/40S complexes with it, and our functional assays confirmed the involvement of eIF3 in the reinitiation process (Fig. 7). As for interactions of the 40S subunit with the 90-nt element, Luttermann and Meyers (2007) have suggested that the critical conserved 5′..UGGGA..3′ core sequence of Motif 1 base-pairs with the apical tetraloop (5′..UCCC..3′) of helix 26 in 18S rRNA. However, as with all such suggestions relating to mammalian ribosomes, this cannot be rigorously tested by making mutations in the 18S rRNA motif and compensatory mutations in the mRNA. In fact, our results show that mutation of the central residues of Motif 1 has a negative effect on the interaction of both eIF3 and 40S subunits with this element (Fig. 6B–D), which is more suggestive of a major structural perturbation than the simple loss of primary sequence complementarity.
Our results fit very nicely with the major new insights into the mechanism of disassembly of post-termination mammalian ribosomes reported by Pisarev et al. (2007), which had hitherto been very much of a mystery, largely because there is no counterpart of prokaryotic RRF in the cytoplasm of eukaryotes. It has been amply demonstrated that although the combination of eRF1, eRF3, and GTP is sufficient to promote release of the completed nascent protein chain, it does not promote disassembly of the post-termination ribosome and its dissociation from the mRNA. Rather, following peptide chain release, initiation factors eIF1, eIF1A, and eIF3 are sufficient and necessary to achieve complete disassembly via the following sequence of events (Pisarev et al. 2007). First, eIF3 promotes dissociation of the 60S subunit, leaving a eIF3/40S complex still bound to the mRNA with a deacylated tRNA in the P-site. Ejection of this deacylated tRNA is promoted by eIF1, and, finally, the loosely associated eIF3j subunit promotes release of the eIF3/40S complex from the mRNA. Gratifyingly, this sequence of events is identical to that operating in prokaryotic systems (Peske et al. 2005), with eIF1 fulfilling the deacylated tRNA ejection role of IF3, in much the same way as the two factors can substitute for each other’s initiator tRNA surveillance function in initiation (Lomakin et al. 2006).
This major breakthrough provides a solid framework for considering possible reinitiation mechanisms on FCV subgenomic mRNA. One possibility is that disassembly of the post-termination ribosome at the ORF2 stop codon proceeds in the normal way up to and including the stage of dissociation of all eIF3/40S complexes from the mRNA, but proximity effects allow some of them to be (re)captured by interaction with the 3′-terminal 90-nt segment of ORF2. The problem with this model is that such a proximity effect would also be expected to allow reinitiation when the ORF2 stop codon lies immediately upstream of the 90-nt segment, or even within it, which is not the case (Fig. 4B,C). In addition, if the 90-nt segment can (re)capture a eIF3/40S complex that has dissociated from the mRNA stop codon and started to diffuse away, it should also have at least some potential for de novo capture of an eIF3/40S complex from the free pool, and to the extent that this occurs, the element should function as an IRES, promoting initiation of ORF3* translation from zero time, which was not observed (Fig. 2A).
We therefore favor models in which those post-termination eIF3/40S complexes that reinitiate translation do not actually dissociate from the mRNA, but instead get transferred directly a very short distance from the stop codon to a “standby” site on the 90-nt segment, where binding is stabilized by interactions of eIF3 and the 40S subunits with this element, and where the position and orientation of the 40S subunit allow the restart codon to enter the ribosomal P-site. There are two variants of this model: In one, all the post-termination ribosomes at the ORF2 stop codon are disassembled by eIF3 entirely recruited from the free pool (just as would happen at any termination codon), but only 10%–15% of the resulting eIF3/40S complexes are successfully transferred from the stop codon to the standby site; in the alternative variant, those ribosomes whose 40S subunits are destined to reinitiate translation are disassembled by eIF3 that binds initially to the 90-nt element and remains so bound while it promotes post-termination ribosome disassembly (while the majority of post-termination ribosomes are disassembled by eIF3 recruited from the free pool). We favor the second variant because it provides a more rational explanation for why supplementary eIF3 and the consequent increase in the free eIF3 pool increases the efficiency of reinitiation, and why this increase is proportionately greater for the 75-nt truncation mutant, which shows reduced affinity for eIF3 and reduced reinitiation efficiency, as compared with the construct that retains the terminal 87 nt of ORF2 (Fig. 7B). In contrast, in the first variant, an increase (or decrease) in the free eIF3 pool will probably increase (decrease) the rate of loading eIF3 on to the post-termination ribosome, but, once loaded, the expectation is that the disassembly of the post-termination ribosome will proceed at the same rate and by the same route as in the control system, and therefore supplementary eIF3 would not be expected to stimulate reinitiation, nor would eIF3 depletion be expected to result in the reduced reinitiation efficiency that was, in fact, observed (Fig. 7D).
Because disassembly of post-termination ribosomes requires eIF3, inevitably leading to formation of eIF3/40S complexes, it seems likely that all such reinitiation events will involve interaction of the eIF3/40S complex with the required cis-acting element. However, this does not rule out the possibility of additional protein factors playing a role in stabilizing the interaction of the eIF3/40S complex with the cis-acting RNA element in some cases, and, indeed, at this stage we cannot definitively rule out such additional protein/RNA interactions in the FCV system. One obvious candidate for an additional stabilizing interaction would be if eIF4G bound to the cis-acting element and captured the post-termination eIF3/40S complex via the well-known eIF3–eIF4G interaction (Hinnebusch 2006), although it is clear that in the particular case of FCV subgenomic mRNA, reinitiation is completely independent of eIF4G (Fig. 2C). What is important to note, however, is that all interactions involved in the capture and retention of the post-termination 40S subunit by the cis-acting mRNA element would have to be quite weak if this element is to function specifically in promoting a reinitiation event following termination at a nearby stop codon, rather than acting as an IRES capable of promoting internal initiation by recruitment of ribosomes from the free pool.
Materials and methods
Plasmid constructs
The construction of the three starting plasmids pSG-2/3*, pCSFV-2/3*, and pCrPV-2/3*, and the mutant derivatives is described in the Supplemental Material. Unless otherwise stated, they were all linearized with XmaI prior to transcription.
In vitro transcription and translation reactions
Capped and uncapped RNAs were synthesized using bacteriophage T7 RNA polymerase, and the RNA product was isolated and quantitated exactly as described by Dasso and Jackson (1989).
In vitro translation reactions were carried out in nuclease-treated RRL as described previously (Kaminski et al. 1990). In brief, the reactions contained 50% (by volume) nuclease-treated reticulocyte lysate (Promega), with 500 μCi/mL [35S]methionine (GE Healthcare, Amersham, SJ1515; >1000 Ci/mmol), and incubation was for 60 min at 30°C unless otherwise stated. Added KCl was at 100 mM for capped transcripts of pSG-2/3*, or 120 mM for uncapped CSFV-2/3* and CrPV-2/3* mRNAs. The standard concentration of mRNA was 50 μg/mL, except where stated otherwise. Translation products were separated by 15% polyacrylamide gel electrophoresis and visualized by autoradiography with Biomax (Kodak) or Hyperfilm βmax (GE Healthcare). Quantitation was carried out by densitometry of the autoradiograms using Phoretix software with several different exposures to ensure that measurements were made within the linear response range of the film, or by PhosphorImaging and volumetric analyses using Imagequant software (Molecular Dynamics).
The preparation of eIF4G-depleted reticulocyte lysate was carried out exactly as described by Ali et al. (2001). The same procedure was used in attempts to deplete eIF3, except that the affinity substrate was a poly(A)-tailed mutated CSFV IRES (CCA331–333 → GGU) incapable of supporting translation but unaffected in eIF3 binding (Supplementary Fig. S4; Kolupaeva et al. 2000). Recombinant His-tagged R362 → Q dominant-negative eIF4A was purified as described by Pestova et al. (1998). HeLa cell eIF3 and 40S ribosomal subunits were purified as described by Siridechadilok et al. (2005).
UV-cross-linking assays
The preparation of HeLa cell HS-S100 (high-salt S100 extract) and its use in UV-cross-linking assays was as described in Hunt and Jackson (1999). E. coli rRNA (Sigma) was present throughout at 100 μg/mL to suppress nonspecific interactions with the probe.
Acknowledgments
We thank Tatyana Pestova and Christopher Hellen for communicating their results prior to publication, and for initiation factor expression constructs; John Hershey and Simon Morley for anti-eIF3 antibodies; T.D.K. Brown for the FCV F9 strain cDNA; and Jenny Reed for technical support. The major part of this work was supported by a Wellcome Trust Programme Grant (062348), and it was completed under a BBSRC Project Grant (BB/D008190/1).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.439507
References
- Adhin M.R., van Duin J., van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- Ahmadian G., Randhawa J.S., Easton A.J., Randhawa J.S., Easton A.J., Easton A.J. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 2000;19:2681–2689. doi: 10.1093/emboj/19.11.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I.K., McKendrick L., Morley S.J., Jackson R.J., McKendrick L., Morley S.J., Jackson R.J., Morley S.J., Jackson R.J., Jackson R.J. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke I.N., Lambden P.R., Lambden P.R. Organization and expression of calicivirus genes. J. Infect. Dis. 2000;181:S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- Dasso M.C., Jackson R.J., Jackson R.J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S.P., Ali I.K., Kaminski A., Digard P., Jackson R.J., Ali I.K., Kaminski A., Digard P., Jackson R.J., Kaminski A., Digard P., Jackson R.J., Digard P., Jackson R.J., Jackson R.J. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA. 2002;8:1558–1571. [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L., Natoni A., Labrie L., Laliberte J.F., Roberts L., Labrie L., Laliberte J.F., Roberts L., Laliberte J.F., Roberts L., Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.S., Easton A.J., Easton A.J. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 2005;280:21972–21980. doi: 10.1074/jbc.M502276200. [DOI] [PubMed] [Google Scholar]
- Herbert T.P., Brierley I., Brown T.D.K., Brierley I., Brown T.D.K., Brown T.D.K. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- Herbert T.P., Brierley I., Brown T.D.K., Brierley I., Brown T.D.K., Brown T.D.K. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Horvath C.M., Williams M.A., Lamb R.A., Williams M.A., Lamb R.A., Lamb R.A. Eukaryotic coupled translation of tandem cistrons: Identification of the influenza B virus BM2 polypeptide. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.L., Jackson R.J., Jackson R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M., Steege D.A., Steege D.A. Mutational analysis of an inherently defective translation initiation site. J. Mol. Biol. 1992;224:1039–1054. doi: 10.1016/0022-2836(92)90468-y. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology. 1986;149:114–127. doi: 10.1016/0042-6822(86)90092-9. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Kaminski A., Pöyry T.A.A., Kaminski A., Pöyry T.A.A., Pöyry T.A.A. Coupled termination–reinitiation events in mRNA translation. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 197–223. [Google Scholar]
- Kaminski A., Howell M.T., Jackson R.J., Howell M.T., Jackson R.J., Jackson R.J. Initiation of encephalomyocarditis virus RNA translation: The authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K.K., Matsumoto T., Fujiwara H., Matsumoto T., Fujiwara H., Fujiwara H. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell. Biol. 2005;25:7675–7686. doi: 10.1128/MCB.25.17.7675-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova T.V., Hellen C.U.T., Pestova T.V., Hellen C.U.T., Hellen C.U.T. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Migration of 40 S ribosomal subunits on messenger RNA when initiation is perturbed by lowering magnesium or adding drugs. J. Biol. Chem. 1979;254:4731–4738. [PubMed] [Google Scholar]
- Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M., Shatkin A.J., Shatkin A.J. Migration of 40S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U., Pestova T.V., Shirokikh N.E., Yusupov M.M., Hellen C.U., Pestova T.V., Yusupov M.M., Hellen C.U., Pestova T.V., Hellen C.U., Pestova T.V., Pestova T.V. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C., Meyers G., Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 2003;278:34051–34056. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- Meyers G. Characterization of the sequence element directing translation reinitiation in the RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Methot N., Svitkin Y., Merrick W.C., Sonenberg N., Methot N., Svitkin Y., Merrick W.C., Sonenberg N., Svitkin Y., Merrick W.C., Sonenberg N., Merrick W.C., Sonenberg N., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F., Rodnina M.V., Wintermeyer W., Rodnina M.V., Wintermeyer W., Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Hellen C.U.T., Hellen C.U.T. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U.T., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U.T., Fletcher S.P., Jackson R.J., Hellen C.U.T., Jackson R.J., Hellen C.U.T., Hellen C.U.T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Hellen C.U.T, Pestova T.V., Hellen C.U.T, Pestova T.V., Pestova T.V. Recycling of eukaryotic post-termination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A.A., Kaminski A., Jackson R.J., Kaminski A., Jackson R.J., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J.E., Kaminski A., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J., Kaminski A., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J., Carroll A.R., Rowlands D.J., Jackson R.J., Rowlands D.J., Jackson R.J., Jackson R.J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabhai A., Brenner S., Brenner S. A mutant which reinitiates the polypeptide chain after chain termination. J. Mol. Biol. 1967;27:145–162. doi: 10.1016/0022-2836(67)90357-9. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B., Fraser C.S., Hall R.J., Doudna J.A., Nogales E., Fraser C.S., Hall R.J., Doudna J.A., Nogales E., Hall R.J., Doudna J.A., Nogales E., Doudna J.A., Nogales E., Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Sizova D.V., Kolupaeva V.G., Pestova T.V., Shatsky I.N., Hellen C.U.T., Kolupaeva V.G., Pestova T.V., Shatsky I.N., Hellen C.U.T., Pestova T.V., Shatsky I.N., Hellen C.U.T., Shatsky I.N., Hellen C.U.T., Hellen C.U.T. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S., Green K.Y., Green K.Y. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Green K.Y., Green K.Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology. 2000;277:193–203. doi: 10.1006/viro.2000.0579. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y., Belliot G., Chang K.O., Onwudiwe O., Green K.Y., Chang K.O., Onwudiwe O., Green K.Y., Onwudiwe O., Green K.Y., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steege D.A. 5′-Terminal nucleotide sequence of Escherichia colilactose repressor mRNA: Features of translational initiation and reinitiation sites. Proc. Natl. Acad. Sci. 1977;74:4163–4167. doi: 10.1073/pnas.74.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Pestova T.V., Hellen C.U.T., Sarnow P., Pestova T.V., Hellen C.U.T., Sarnow P., Hellen C.U.T., Sarnow P., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;123:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wirblich C., Thiel H.J., Meyers G., Thiel H.J., Meyers G., Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]