Abstract
Apoptosis is essential for proper development and tissue homeostasis in metazoans. It plays a critical role in generating sexual dimorphism by eliminating structures that are not needed in a specific sex. The molecular mechanisms that regulate sexually dimorphic apoptosis are poorly understood. Here we report the identification of the ceh-30 gene as a key regulator of sex-specific apoptosis in Caenorhabditis elegans. Loss-of-function mutations in ceh-30 cause the ectopic death of male-specific CEM neurons. ceh-30 encodes a BarH homeodomain protein that acts downstream from the terminal sex determination gene tra-1, but upstream of, or in parallel to, the cell-death-initiating gene egl-1 to protect CEM neurons from undergoing apoptosis in males. The second intron of the ceh-30 gene contains two adjacent cis-elements that are binding sites for TRA-1A and a POU-type homeodomain protein UNC-86 and acts as a sensor to regulate proper specification of the CEM cell fate. Surprisingly, the N terminus of CEH-30 but not its homeodomain is critical for CEH-30’s cell death inhibitory activity in CEMs and contains a conserved eh1/FIL domain that is important for the recruitment of the general transcriptional repressor UNC-37/Groucho. Our study suggests that ceh-30 defines a critical checkpoint that integrates the sex determination signal TRA-1 and the cell fate determination and survival signal UNC-86 to control the sex-specific activation of the cell death program in CEMs through the general transcription repressor UNC-37.
[Keywords: C. elegans, apoptosis, sex-specific, unc-37/Groucho, ceh-30, transcriptional regulation]
The development of sexually dimorphic traits, those systematic differences between two sexes of an organism, is an important process in metazoan development. In many cases, sex-specific apoptosis plays an essential role in shaping the sexual dimorphism of animals (Roberts et al. 1999). For example, during male development in mammals, a Mullerian-inhibiting substance triggers apoptosis and regression of the Mullerian ducts that would develop into the female reproductive system (Price et al. 1977). Alternately, the Wolffian ducts, which would mature into the male-specific reproductive structures, undergo apoptosis during the development of a female (Yin et al. 2006). Additional instances in which apoptosis regulates sexually dimorphic development include Bax-dependant sexual differentiation of cell numbers in two regions of the mouse forebrain, loss of lactotrophs in male rats, and mammary epithelial cell destruction in male mice (Dunbar et al. 1999; Aoki et al. 2001; Forger et al. 2004). Apoptosis also plays important roles in some diseases with sexually dimorphic outcomes or treatment efficacies, such as susceptibility to diabetes and renal injury (Casteels et al. 1998; Blush et al. 2004). Studying the regulation of apoptosis leading to these sex-specific differences is important for our understanding of sexual dimorphism and its implications on the development and treatment of related human diseases.
Caenorhabditis elegans has two sexes, males and self-reproducing hermaphrodites, which also exhibit sexually dimorphic apoptosis. A pair of bilaterally symmetric motor neurons named HSNs (hermaphrodite-specific neurons) control egg-laying in hermaphrodites and undergo apoptosis in males, where they are not needed (Sulston and Horvitz 1977; Sulston et al. 1983). In contrast, four male-specific chemosensory neurons (CEMs; cephalic companion neurons), which are thought to mediate chemotaxis of the male toward the hermaphrodite during courtship behavior, undergo apoptosis in hermaphrodites (Sulston and Horvitz 1977; Sulston et al. 1983). The sex-specific deaths of HSNs and CEMs provide a simple model for studying the molecular mechanisms that control sexually dimorphic apoptosis.
During the development of C. elegans hermaphrodites, 131 cells die in an invariant pattern (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). Genetic analysis in C. elegans has led to the identification of four genes (egl-1, ced-9, ced-4, and ced-3) that act in a sequential manner to induce apoptosis (Ellis and Horvitz 1986; Hengartner et al. 1992; Conradt and Horvitz 1998). Biochemical studies indicate that EGL-1, a BH3-only protein, induces cell death by binding to CED-9, a cell death inhibitor and human Bcl-2 homolog, causing the release of CED-4 from the CED-4/CED-9 complex tethered to the outer mitochondrial membrane and the subsequent CED-4-mediated activation of the cysteine protease CED-3 (the cell death executioner) (Hengartner et al. 1992; Conradt and Horvitz 1998; Parrish et al. 2000; Yan et al. 2005). How apoptosis is activated in specific cells in response to death signals is poorly understood.
The sexual development of C. elegans soma is controlled by a negative regulatory cascade, initiated by HER-1, a secreted protein, which promotes male development by inhibiting the activity of TRA-2, a transmembrane receptor (Kuwabara et al. 1992; Perry et al. 1993). TRA-2 acts by inhibiting the activities of three interacting cytoplasmic proteins—FEM-1, FEM-2, and FEM-3—which promote male development by inhibiting the activity of TRA-1A, the terminal global regulator of somatic sex determination, through an unknown mechanism (Doniach and Hodgkin 1984; Kimble et al. 1984; Hodgkin 1988; Chin-Sang and Spence 1996; Meyer 1997; Mehra et al. 1999). TRA-1A, a zinc-finger protein, promotes female development by transcriptionally activating female-specific genes and/or by repressing male-specific genes (Zarkower and Hodgkin 1992, 1993). One TRA-1A target is the death-activating gene egl-1, which contains a TRA-1A-binding site 5.6 kb downstream from its ORF (Conradt and Horvitz 1999). Several gain-of-function (gf) mutations in egl-1 disrupt this TRA-1-binding site and cause ectopic expression of egl-1 in hermaphrodite HSNs and inappropriate HSN death, suggesting that TRA-1A represses the expression of egl-1 in hermaphrodite HSNs (Conradt and Horvitz 1999). However, as a general sex determination factor that affects many somatic cells in C. elegans (Zarkower and Hodgkin 1992), TRA-1A does not affect most of the somatic cell deaths and needs to act with HSN-specific factors to control the sexually dimorphic death of HSNs.
Far less is known about what controls the life versus death decision of the male-specific CEM neurons. We undertook a genetic screen to search for mutations affecting CEM cell fates and sex-specific apoptosis. Here, we report the characterization of ceh-30, which is required specifically for the survival of CEMs in males and encodes a BarH homeodomain protein. We show that TRA-1A and a POU-type homeodomain protein, UNC-86, act concertedly to regulate sex-specific expression of ceh-30 in CEMs, leading to proper CEM cell death specification. CEH-30 then acts through a conserved eh1/FIL domain at its N terminus to recruit the general transcriptional repressor Groucho/UNC-37, resulting in the formation of a transcriptional repressosome that suppresses programmed cell death. Thus ceh-30 represents a critical link between the sex determination pathway and the programmed cell death pathway that controls sex-specific apoptosis in C. elegans.
Results
A genetic screen to identify genes that regulate CEM cell death specification
To identify factors that control sex-specific CEM death, we carried out a green fluorescent protein (GFP)-based screen using a CEM-specific reporter construct (Ppkd-2gfp) to isolate mutations that alter the sexually dimorphic cell fates of CEMs (Barr and Sternberg 1999). The pkd-2 promoter drives GFP expression in four CEM neurons and some ray neurons in the tail of male animals; no GFP expression is observed in hermaphrodites (data not shown). We generated an integrated transgenic array containing Ppkd-2gfp (smIs23) in him-5(e1490) animals, which produce a high frequency of male progeny as a result of increased meiotic nondisjunction (Goldstein 1986). We then performed ethyl methane sulfonate (EMS) mutagenesis on the resulting strain, smIs23; him-5(e1490), and screened for mutations that cause ectopic CEM loss in males or improper CEM survival in hermaphrodites (see Materials and Methods).
We screened 8500 haploid genomes and isolated several mutations that affect the cell fate specification of CEMs and/or HSNs (E. Peden, E. Kimberly, and D. Xue, unpubl.), including the sm130 mutation and a mutation (sm117) that affects a previously identified gene, unc-86.
sm130 is a X-linked mutation that causes ectopic CEM death in males
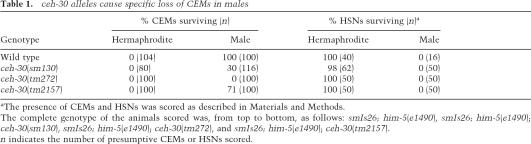
In him-5(e1490) animals carrying smIs26, another Ppkd-2gfpcontaining integrated array that also harbors Ptph-1gfp (a HSN reporter) (Sze et al. 2000), 100% of CEMs survive in males (Table 1). In contrast, only 30% of CEMs are present in smIs26; him-5(e1490); ceh-30(sm130) males. The loss of CEMs in sm130 males can be suppressed by a strong loss-of-function mutation in either the egl-1 gene (n3082) or the ced-3 gene (n717), suggesting that these CEMs die by programmed cell death rather than adopting a non-CEM cell fate (Table 2). sm130 does not affect the sex-specific death of HSNs (Table 1) or the deaths and sexual differentiation of other somatic cells (data not shown). Therefore, sm130 appears to affect a gene specifically regulating CEM cell death.
Table 1.
ceh-30 alleles cause specific loss of CEMs in males
aThe presence of CEMs and HSNs was scored as described in Materials and Methods.
The complete genotype of the animals scored was, from top to bottom, as follows: smIs26; him-5(e1490), smIs26; him-5(e1490); ceh-30(sm130), smIs26; him-5(e1490); ceh-30(tm272), and smIs26; him-5(e1490); ceh-30(tm2157).
n indicates the number of presumptive CEMs or HSNs scored.
Table 2.
ceh-30 acts downstream from tra-1 and upstream of, or in parallel to, egl-1 to regulate sex-specific CEM death
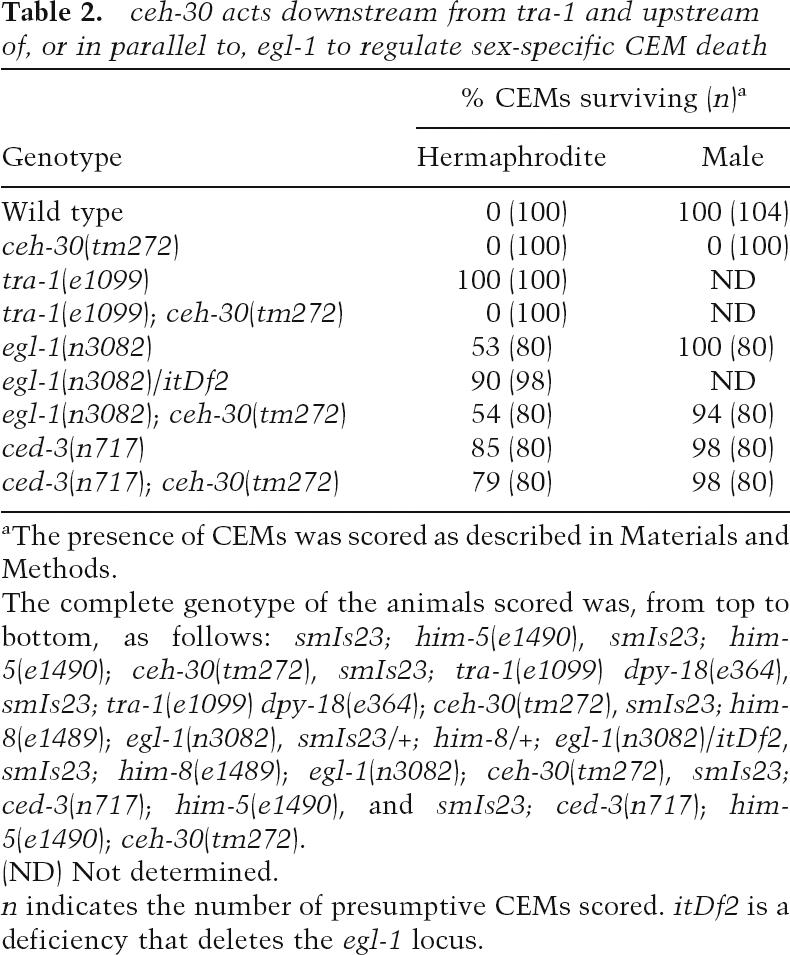
aThe presence of CEMs was scored as described in Materials and Methods.
The complete genotype of the animals scored was, from top to bottom, as follows: smIs23; him-5(e1490), smIs23; him-5(e1490); ceh-30(tm272), smIs23; tra-1(e1099) dpy-18(e364), smIs23; tra-1(e1099) dpy-18(e364); ceh-30(tm272), smIs23; him-8(e1489); egl-1(n3082), smIs23/+; him-8/+; egl-1(n3082)/itDf2, smIs23; him-8(e1489); egl-1(n3082); ceh-30(tm272), smIs23; ced-3(n717); him-5(e1490), and smIs23; ced-3(n717); him-5(e1490); ceh-30(tm272).
(ND) Not determined.
n indicates the number of presumptive CEMs scored. itDf2 is a deficiency that deletes the egl-1 locus.
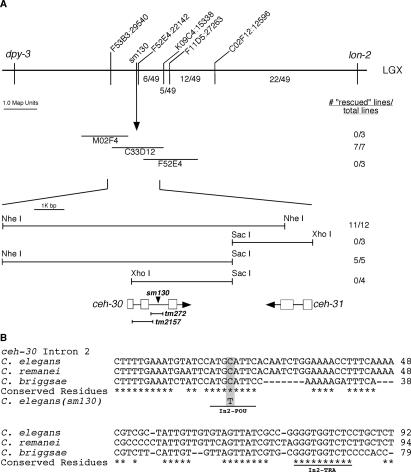
Using the phenotype of ectopic male CEM death, sm130 was mapped to Linkage Group X (LGX) between two genetic markers, lon-2 and unc-2, at ∼3.03 Mb or −12.7 cM, which was covered by three overlapping Cosmids, M02F4, C33D12, and F52E4 (see Fig. 1A; Materials and Methods).
Figure 1.
Mapping and rescue of the sm130 mutant. (A, top panel) The genetic markers and SNPs used to map the sm130 mutation are indicated above the horizontal bar. The numbers below the bar indicate the fraction of 49 recombinants between lon-2 and sm130 that occurred between particular SNP markers. (Middle panel) Cosmids tested for rescue. Transgenic animals carrying the indicated Cosmid DNA as extrachromosomal arrays were generated and scored for rescue of the ectopic CEM death defect in the sm130 mutant as described in Materials and Methods. The number of rescued lines versus total lines generated is indicated to the right. (Bottom panel) Partial restriction map of the rescuing Cosmid C33D12 and the sm130 rescue data observed for several subclones of C33D12. ORFs in this region are indicated with boxes representing exons and lines representing intronic sequences. The position of the sm130 mutation is indicated with an arrowhead. Two deletion alleles (tm272 and tm2157) of ceh-30 and the ceh-30 regions removed by these mutations are represented below the ceh-30 ORF. (B) Alignment of the ceh-30 intron 2 DNA sequences from three related nematode species (C. elegans, C. briggsae, and C. remanei). Conserved residues are indicated by asterisks. The sm130 lesion is highlighted, and two conserved binding sites for POU-type homeodomain proteins (In2-POU) and TRA-1A (In2-TRA) are underlined.
ceh-30 encodes a BarH homeodomain protein that is essential for CEM survival in males
To clone the gene affected by sm130, germline transformations of smIs23; him-5(e1490); sm130 animals were carried out by injecting three individual Cosmids (M02F4, C33D12, and F52E4) spanning the locus defined by the sm130 mutation. Only Cosmid C33D12 showed rescue of the ectopic male CEM death phenotype. Further analysis of C33D12 subclones indicates that a single ORF, ceh-30, which encodes a BarH homeodomain-containing protein, is responsible for the rescuing activity (Fig. 1A).
Sequencing of genomic DNA from the sm130 mutant revealed a single nucleotide substitution (a C-to-T transition) 336 base pairs (bp) into the second intron of ceh-30 (Fig. 1A). We also obtained two deletion alleles of ceh-30, tm272, and tm2157, which remove 425 bp and 718 bp from the ceh-30 coding region, respectively (Fig. 1A). ceh-30(tm272) deletes part of the second intron of ceh-30, including the nucleotide mutated by sm130, and causes 100% CEM death in mutant males (Table 1). ceh-30(tm2157) removes part of exon 1 and all of intron 1 and exon 2 but leaves most of intron 2 intact, including the region affected by sm130 (Fig. 1A). Interestingly, tm2157 only causes a weak reduction of male CEM survival (29%), albeit removing most of the CEH-30 protein including its entire homeodomain (see below). These results underscore the critical role of the ceh-30 second intron in protecting CEMs from undergoing apoptosis in males. Like sm130, ceh-30(tm272) and ceh-30(tm2157) do not affect the death of HSNs or the death and sex differentiation of other somatic cells (Table 1; data not shown).
ceh-30 acts downstream from tra-1 and upstream of, or in parallel to, egl-1 to regulate sex-specific CEM death
To understand how ceh-30 acts to control sexually dimorphic apoptosis of CEMs, we carried out genetic epistasis analysis to determine the relationships between ceh-30 and key regulators of sex determination and apoptosis. In particular, we focused on the terminal sex determination gene tra-1, the cell death initiator egl-1, and the cell death executor ced-3 (Ellis and Horvitz 1986; Zarkower and Hodgkin 1992; Conradt and Horvitz 1999).
tra-1 acts at the terminal step of the C. elegans sex determination pathway to control the sexual fates of somatic cells, including the fates of HSN and CEM neurons. Hermaphrodite XX animals homozygous for the null tra-1(e1099) mutation develop as low-fertility males with 100% inappropriately surviving CEMs (Table 2; Hodgkin and Brenner 1977). However, in smIs23; tra-1(e1099); ceh-30(tm272) XX animals, CEMs are never present. Given that tra-1 is the terminal sex determination gene, this result suggests that ceh-30 acts downstream from, or in parallel to, tra-1 as well as the entire sex determination pathway to control the sex-specific death of CEMs.
egl-1 is proposed to initiate the activation of all somatic cell deaths in C. elegans (Conradt and Horvitz 1998). In smIs23; egl-1(n3082); ceh-30(tm272) males, the ectopic CEM death phenotype caused by ceh-30(tm272) is almost completely suppressed by egl-1(n3082), suggesting that ceh-30 acts upstream of, or in parallel to, egl-1 to control sex-specific CEM deaths. Intriguingly, ceh-30 can also regulate CEM death in the absence of either egl-1 or ced-9, suggesting that ceh-30 can act in parallel to egl-1 and ced-9 to control sex-specific CEM death (H.T. Schwartz and H.R. Horvitz, pers. comm.). Like egl-1(n3082), ced-3(n717) also potently suppresses ectopic CEM death in ceh-30(tm272) males (Table 2). Taken together, these results suggest that ceh-30 acts downstream from the sex determination pathway but upstream of, or in parallel to, the C. elegans programmed cell death pathway to control sexually dimorphic deaths of CEM neurons and may serve as a critical mediator between these two globally acting pathways to regulate cell-type and sex-specific deaths.
ceh-30(sm130) and ceh-30(tm272) affect a conserved binding site for POU-type homeodomain proteins
Two ceh-30 alleles, sm130 and tm272, alter or remove sequences within intron 2 of the ceh-30 gene. The C-to-T transition caused by sm130 falls within a conserved sequence motif (CATGCATTC) in intron 2 of the ceh-30 genes from three different nematode species (Caenorhabditis briggsae, Caenorhabditis remanei, and Caenorhabditis elegans) (Fig. 1B). By searching the Transcription Factor Database (TFD), we found that this sequence shares high sequence identity with the binding sites of the mammalian POU-type homeodomain proteins GHF/Pit1 (ATGCATTC) and Brn-2/N-Oct-3 [ATG(A/C)AT(A/T)] (Argenton et al. 1996; Rhee et al. 1998). In addition, the C. elegans POU-type homeodomain protein UNC-86 has been shown to recognize a similar sequence, ATG(A/C)AT (Finney et al. 1988; Xue et al. 1992). These observations suggest that sm130 might affect the binding site of a POU-type homeodomain protein and that this site, which we call In2-POU, is important for ceh-30’s function in preventing CEM death in males.
There are several additional conserved sequence motifs in intron 2 downstream from In2-POU (Fig. 1B). One of them, located 55 bp downstream, resembles a conserved TRA-1A-binding site (Zarkower and Hodgkin 1993). This site, which we name In2-TRA, is altered in several ceh-30 gain-of-function mutants where CEMs survive inappropriately in hermaphrodites (H.T. Schwartz and H.R. Horvitz, pers. comm.). These findings suggest that intron 2 of ceh-30 is critical for the regulation of sex-specific CEM death. Indeed, the tm272 deletion that removes both In2-POU and In2-TRA motifs results in a complete absence of CEMs in males.
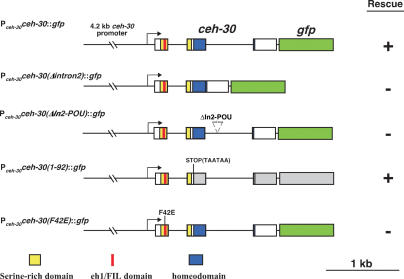
To verify the importance of ceh-30 intron 2 and In2-POU in regulating the sexually dimorphic cell fate of CEMs, we generated a ceh-30 translational GFP fusion (Pceh-30ceh-30∷gfp) that contains a 4.2-kb ceh-30 promoter as well as the entire ceh-30 ORF and found that this construct rescues the CEM defect of the ceh-30(tm272) mutant (Fig. 2). In contrast, a Pceh-30ceh-30(Δintron2)∷gfp construct that lacks all of intron 2 or a Pceh-30ceh-30(ΔIn2-POU)∷gfp construct in which the 8-bp In2-POU site (ATGCATTC) is deleted failed to rescue the ceh-30(tm272) mutant (Fig. 2), providing further evidence that intron 2 and the In2-POU site of ceh-30 are critical for its apoptosis-inhibitory activity in male CEMs.
Figure 2.
The N terminus of CEH-30 but not its homeodomain is required for its activity to protect male CEMs from cell death. Graphical presentations of five ceh-30 constructs are shown. Boxes indicate ceh-30 exons or the coding region for GFP (in green). The region encoding the homeodomain is highlighted with blue, the region encoding the SRD is highlighted with yellow, and the region encoding the eh1/FIL motif is highlighted with red. Transgenic smIs26; him-5(e1490); ceh-30(tm272) animals carrying the indicated DNA construct as extrachromosomal arrays were scored for rescue of the ectopic CEM death defect as described in Materials and Methods. For each construct, at least three independent transgenic lines were scored and found to have similar results. (+) Rescue; (−) failure to rescue.
unc-86 is required for CEM cell fate specification independent of ced-3
Our CEM cell fate mutant screen also identified an allele of unc-86 (sm117) that results in almost complete absence of GFP expression at the CEM positions in smIs23 males (Table 3). However, very few GFP-expressing cells are seen at the CEM positions in smIs23; unc-86(sm117); ced-3(n717) males, indicating that unc-86 is required for CEM cell fate specification at a step prior to the life versus death decision of CEMs. We also observed almost complete loss of GFP expression at the CEM positions in several other unc-86 loss-of-function mutants (Table 3). UNC-86 is expressed in 47 neurons during C. elegans development, including male CEMs and hermaphrodite HSNs (Finney and Ruvkun 1990), and is critical for the proper differentiation of these UNC-86-expressing neurons, including the differentiation and survival of CEM neurons in males (Finney et al. 1988; Shaham and Bargmann 2002). Interestingly, the mouse ortholog of UNC-86, Brn3c, has been shown to act genetically upstream of the mouse BarH homeodomain protein Barhl1 to promote terminal differentiation and survival of mechanosensory hair cells (Li et al. 2002), suggesting a conserved regulatory relationship between these two types of homeodomain proteins. Therefore, unc-86 is an excellent candidate for the transcriptional activator of ceh-30 through the In2-POU cis-element.
Table 3.
unc-86 affects CEM cell fate
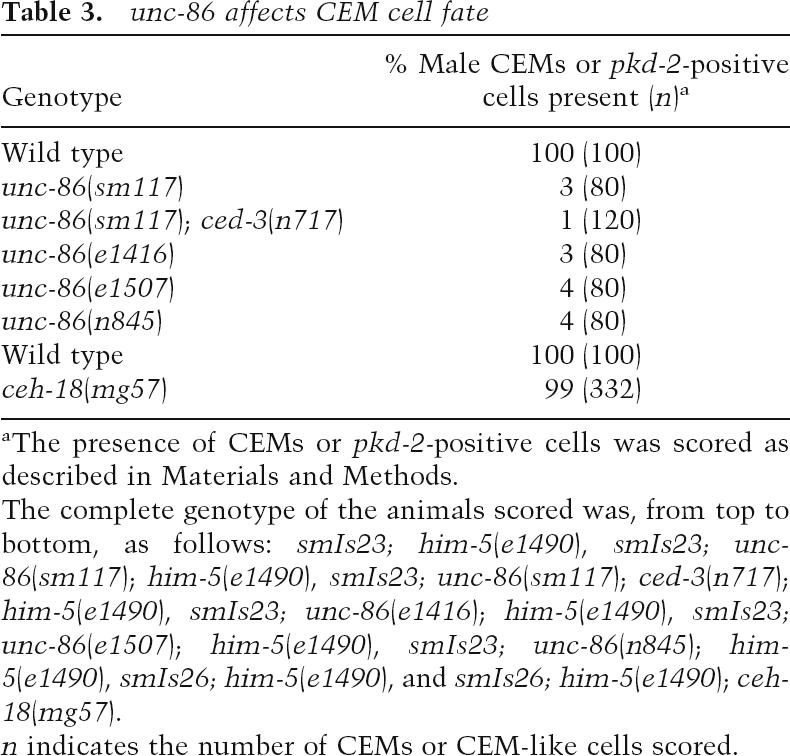
aThe presence of CEMs or pkd-2-positive cells was scored as described in Materials and Methods.
The complete genotype of the animals scored was, from top to bottom, as follows: smIs23; him-5(e1490), smIs23; unc-86(sm117); him-5(e1490), smIs23; unc-86(sm117); ced-3(n717); him-5(e1490), smIs23; unc-86(e1416); him-5(e1490), smIs23; unc-86(e1507); him-5(e1490), smIs23; unc-86(n845); him-5(e1490), smIs26; him-5(e1490), and smIs26; him-5(e1490); ceh-18(mg57).
n indicates the number of CEMs or CEM-like cells scored.
In addition to unc-86, there are only two other genes in the C. elegans genome that encode POU-type homeodomain proteins (ceh-18 and ceh-6). The ceh-18-null allele (mg57) does not cause any defect in CEM cell fate specification (Table 3; Greenstein et al. 1994). Animals homozygous for the only available ceh-6 allele (mg60) are lethal at the embryonic and L1 larval stages, making analysis of the CEM cell fate impossible. Analysis of CEH-6 expression using anti-CEH-6 antibody and reporter constructs does not identify CEH-6 expression in CEMs or HSNs (Burglin and Ruvkun 2001). These data suggest that ceh-18 does not play a role in CEM cell fate specification, while a role for ceh-6 in CEM fate determination is unlikely but cannot be ruled out.
UNC-86 and TRA-1A bind intron 2 of ceh-30 in vitro
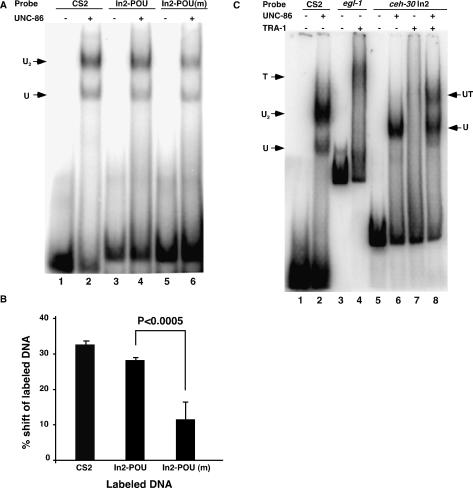
We next used the gel mobility shift assay to investigate the possibility that UNC-86 binds the In2-POU site. We found that full-length UNC-86 protein tagged with glutathione S-transferase (GST) at its N terminus (GST∷UNC-86) can bind to a known UNC-86-binding site (CS2) from the promoter of the mec-3 gene (Xue et al. 1993) as well as a 33-bp DNA fragment containing either the In2-POU site or the altered In2-POU site [In2-POU(m)] found in the sm130 mutant (Fig. 3A). Quantification of the amount of DNA shifted due to GST∷UNC-86 binding in four independent experiments reveals that binding to In2-POU(m) (12% DNA shifted) is significantly weaker than binding to In2-POU (28% DNA shifted; P-value < 0.0005) (Fig. 3B). This decrease in binding efficiency of GST∷UNC-86 to the In2-POU(m) site is consistent with the reduced, but not abolished, ceh-30 activity observed in the ceh-30(sm130) mutant (Table 1).
Figure 3.
Both UNC-86 and TRA-1A bind to the intron 2 sequence of ceh-30 in vitro. (A) Binding of GST∷UNC-86 to a POU-type homeodomain-binding site (In2-POU) found in intron 2 of ceh-30 and an altered site [In2-POU(m)] derived from the sm130 mutant. Twenty-five nanograms of recombinant GST∷UNC-86 (lanes 2,4,6) or binding buffer alone (lanes 1,3,5) were incubated with labeled oligonucleotides derived from a known UNC-86-binding site (CS2; lanes 1,2), In2-POU (lanes 3,4), or In2-POU(m) (lanes 5,6) and resolved on an 8% native polyacrylamide gel. (U) The gel shift species containing the UNC-86 monomer; (U2) the gel shift species containing the UNC-86 homodimer (Xue et al. 1993). (B) The sm130 lesion reduces the binding of GST∷UNC-86 to In2-POU. The fraction of labeled probe shifted by GST∷UNC-86 was quantified from four independent experiments as described in Materials and Methods. (C) UNC-86 stabilizes the binding of TRA-1A to the intron 2 sequence of ceh-30. Twenty-five nanograms of recombinant GST∷UNC-86 and 5 ng of TRA-1∷His6 either alone or together were incubated with labeled CS2 (lanes 1,2), 257-bp DNA fragment containing the TRA-1A-binding site from the egl-1 gene (egl-1, lanes 3,4), or 91-bp intron 2 DNA fragment from ceh-30 containing both In2-POU and In2-TRA (ceh-30 In2, lanes 5–8). The reactions were resolved on a 4% native polyacrylamide gel. (T) The gel shift species containing the TRA-1 protein; (UT) the gel shift species containing both UNC-86 and TRA-1A.
Given that tra-1 acts upstream of ceh-30 and that a potential TRA-1A-binding site (In2-TRA) is located 55 bp downstream from the In2-POU site, we investigated if TRA-1A binds to this site and if the binding of TRA-1A to this site affects the binding of UNC-86 to the neighboring In2-POU site or vice versa. As shown in Figure 3C, the full-length TRA-1A protein tagged with hexa-histidine (TRA-1∷His6) can bind a 257-bp DNA fragment containing the critical TRA-1A-binding site from the egl-1 gene (Fig. 3C, lanes 3,4; Conradt and Horvitz 1999). When we included a 91-bp DNA fragment carrying both the In2-POU site and the In2-TRA site in the binding reactions, we found that both GST∷UNC-86 and TRA-1∷His6 were capable of binding to this DNA fragment, although the binding of TRA-1A to this DNA fragment was less stable, displaying a smear gel-shift pattern reflective of the formation of unstable DNA/protein complexes (Fig. 3C, lanes 5–7). Interestingly, addition of GST∷UNC-86 to the TRA-1A-binding reaction stabilizes the binding of TRA-1∷His6 to this 91-bp ceh-30 intronic sequence, resulting in the formation of a discrete TRA-1/UNC-86 supershifted band at the expense of the UNC-86/DNA complexes (Fig. 3C, lane 8). We confirmed the presence of both TRA-1 and UNC-86 in this DNA complex by an antibody supershift experiment (Supplementary Fig. S1). The observation that UNC-86 can stabilize the binding of TRA-1A to the intron 2 sequence suggests that these two proteins interact on this important DNA element to regulate the expression of ceh-30 and thus the proper sex-specific death of CEM neurons.
CEH-30 is widely expressed during embryonic development including in male CEM neurons
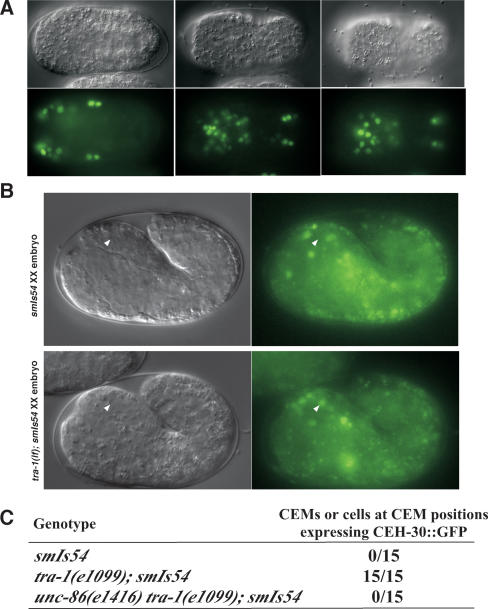
We next determined the expression pattern of ceh-30 during C. elegans development using an integrated transgene (smIs54) carrying Pceh-30ceh-30∷gfp that rescues the CEM defect of the ceh-30(tm272) mutant (Fig. 2). The CEH-30∷GFP fusion is expressed widely during embryonic development, and the expression is especially robust in the anterior half of the embryos (Fig. 4A). CEH-30∷GFP expression continues into the larval and adult stages, albeit at greatly reduced levels.
Figure 4.
CEH-30 expression patterns in C. elegans embryos. (A) GFP expression directed from an integrated array (smIs54) containing Pceh-30ceh-30∷gfp at ∼360 min post-first cleavage. Three different focal planes of the same embryo are shown with the anterior toward the left and posterior toward the right. (B) Expression of CEH-30∷GFP in a smIs54 XX embryo (∼420 min post-first cleavage, top panel) and a tra-1(e1099); smIs54 XX embryo at the similar stage (bottom panel). The ventral CEM neuron is indicated by a white arrowhead. (C) Frequency of CEH-30∷GFP expression in ventral CEMs of XX embryos at ∼380–430 min after first cleavage. The genotypes of XX embryos are indicated.
We then examined whether CEH-30 is expressed in the CEM neurons by following the expression of CEH-30∷GFP in ventral CEM neurons at ∼400–430 min after first cleavage, which are slightly after their births at ∼320 min (Sulston et al. 1983). In hermaphrodite smIs54 embryos, CEH-30∷GFP was never seen in ventral CEMs (0/15 embryos) (Fig. 4C); this result is consistent with the predicted absence of CEH-30 expression in hermaphrodite CEMs. In contrast, in tra-1(n1099); smIs54 XX embryos, which are completely masculinized and virtually identical to male embryos, CEH-30∷GFP was observed in all ventral CEMs (15/15 embryos) (Fig. 4B,C), suggesting that CEH-30 is expressed in developing male CEM neurons. Given that TRA-1A directly binds to intron 2 of ceh-30 (Fig. 3C) and mutations in In2-TRA of ceh-30 cause improper CEM survival in hermaphrodites (H.T. Schwartz and H.R. Horvitz, pers. comm.), this result suggests that TRA-1 directly suppresses CEH-30 expression in hermaphrodite CEMs. Interestingly, in unc-86(e1416) tra-1(e1099); smIs54 XX masculinized embryos, CEH-30∷GFP was not seen in ventral CEM-like cells (0/15 embryos) (Fig. 4C), suggesting that UNC-86 is important for CEH-30 expression in CEMs. Taken together, these results are consistent with the model that UNC-86 activates and TRA-1 inhibits the expression of CEH-30 in CEMs.
The CEH-30 homeodomain is largely dispensable for CEM survival in males
Homeodomains mediate binding to DNA and are critical for the functions of homeodomain-containing proteins (Hayashi and Scott 1990). One ceh-30 deletion allele, tm2157, removes the last 45 bp of exon 1, the entirety of intron 1 and exon 2, and the first 141 bp of intron 2, but leaves the In2-POU site and the In2-TRA site intact (Fig. 1A). This deletion results in the removal of the CEH-30 homeodomain, and presumably, the synthesis of a truncated CEH-30 protein containing only the first 46 amino acids. Despite the large truncation, ceh-30(tm2157) animals show only a weak reduction (29% reduction) in male CEM survival (Table 1), suggesting that the homeodomain of CEH-30 is largely dispensable for its activity in protecting against CEM cell death in males. To confirm this result, we generated a ceh-30∷gfp fusion construct that carries tandem stop codons at the beginning of the ceh-30 BarH homeodomain-coding region [Pceh-30ceh-30(1–92)∷gfp], resulting in the synthesis of a 92-amino-acid N-terminal fragment of CEH-30. This construct rescues the ceh-30(tm272) defect (Fig. 2), providing further support to the idea that the N terminus of CEH-30 is capable of specifying the CEM life versus death decision in the absence of its homeodomain.
The N-terminal eh1/FIL motif of CEH-30 and UNC-37/Groucho are required to protect against CEM cell death in males
Analysis of the CEH-30 protein sequence reveals only three conserved domain structures: the BarH homeodomain (amino acids 94–154), a serine-rich domain (SRD) (amino acids 29–80), and an engrailed homology domain or FIL (eh1/FIL) protein interaction motif (amino acids 42–49) embedded within the SRD (Fig. 5A). The putative ceh-30 transcript made in the ceh-30(tm2157) mutant would encode a truncated CEH-30 protein with only the first 17 amino acids of the SRD domain (52 amino acids in length) and the first five amino acids of the eh1/FIL motif (FRISD from FRISDILE). The eh1/FIL domain was first identified as a motif required for the association of fly homeodomain proteins with the general transcriptional repressor Groucho (Smith and Jaynes 1996; Choi et al. 1999; Jimenez et al. 1999; Bae et al. 2003).
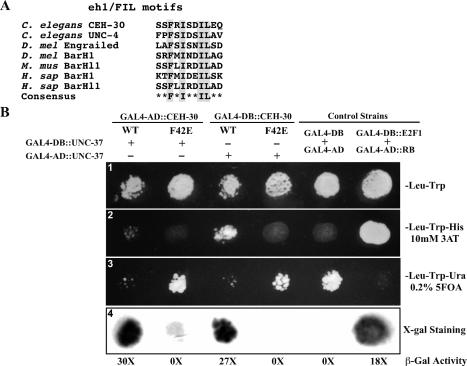
Figure 5.
CEH-30 and UNC-37 interact through the eh1/FIL motif. (A) Alignment of the eh1/FIL motifs in various proteins from C. elegans, Drosophila (D. Mel), mouse (M. Mus), and human (H. sap). Highly conserved amino acid residues are highlighted. (B) Yeast two-hybrid assays. (Panel 1) Yeast transformants expressing CEH-30 and UNC-37 GAL4 fusions as indicated were spotted on synthetic complete medium lacking trytophan and leucine (SC-Leu-Trp). (Panel 2) Growth on the SC-Leu-Trp-His plate with 10 mM 3AT. Growth indicates an interaction between the two tested fusion proteins. (Panel 3) Growth on the SC-Leu-Trp-Ura plate containing 0.2% 5FOA. Poor or no growth on the SC-Leu-Trp-Ura plate with 0.2% 5FOA indicates interaction between two tested fusion proteins. (Panel 4) β-Galactosidase staining was performed on a nitrocellulose membrane as described in Materials and Methods. Dark staining indicates interaction between the two tested fusion proteins. β-Galactosidase activity was also measured by the Chlorophenol red-β-D-galactopyranoside (CPRG) cleavage assay and is indicated below the staining assay. The numbers shown are folds of increase in activity over the yeast strain that expressed GAL4-AD and GAL4-DB. Fulllength CEH-30 (wild type or F42E mutant) and UNC-37 proteins were fused to either the GAL4-AD or the GAL4-DB. GAL4-DB/GAL4-AD and GAL4-DB∷E2F1/GAL4-AD∷RB were used as negative and positive controls for protein interaction, respectively.
We thus tested whether this motif is important for protecting against CEM death in males by substituting Phe 42, a conserved and critical residue in the eh1/FIL motif, with glutamate (Smith and Jaynes 1996; Jennings et al. 2006). A ceh-30∷gfp fusion construct harboring this mutation [Pceh-30ceh-30(F42E)∷gfp] failed to rescue the ceh-30(tm272) mutant, indicating that the eh1/FIL motif of CEH-30 is required to inhibit CEM cell death in males (Fig. 2).
We then examined animals defective in the unc-37 gene, which encodes the C. elegans Groucho ortholog (Pflugrad et al. 1997; Winnier et al. 1999), to determine if unc-37 is involved in CEM cell fate specification. Male animals carrying the only viable allele of unc-37(e262) showed decreased CEM survival (Table 4), whereas the unc-37(e262); ced-3(n2433) double mutant showed normal CEM survival in male animals. We also examined CEM survival in unc-37(e262); ceh-30(lf) double mutants and found that two partial loss-of-function ceh-30 alleles (sm130 and tm2157) either additively or synergistically reduced CEM survival with the unc-37(e262) mutation (Table 4). In particular, unc-37(e262) and ceh-30(tm2157) mutations, which individually only mildly reduce male CEM survival (17% and 29% male CEM death, respectively), can synergistically cause most CEMs to die in males (74% CEM death). These results indicate that unc-37 is important for maintaining CEM survival in males and likely acts with CEH-30 to inhibit male CEM cell death.
Table 4.
unc-37 affects CEM cell fate
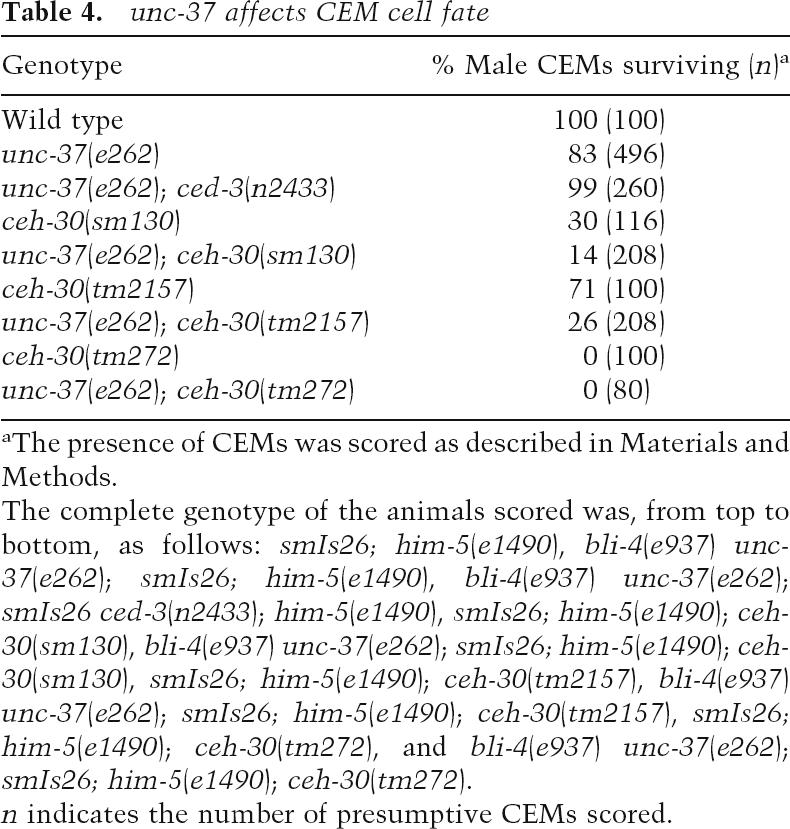
aThe presence of CEMs was scored as described in Materials and Methods.
The complete genotype of the animals scored was, from top to bottom, as follows: smIs26; him-5(e1490), bli-4(e937) unc-37(e262); smIs26; him-5(e1490), bli-4(e937) unc-37(e262); smIs26 ced-3(n2433); him-5(e1490), smIs26; him-5(e1490); ceh-30(sm130), bli-4(e937) unc-37(e262); smIs26; him-5(e1490); ceh-30(sm130), smIs26; him-5(e1490); ceh-30(tm2157), bli-4(e937) unc-37(e262); smIs26; him-5(e1490); ceh-30(tm2157), smIs26; him-5(e1490); ceh-30(tm272), and bli-4(e937) unc-37(e262); smIs26; him-5(e1490); ceh-30(tm272).
n indicates the number of presumptive CEMs scored.
Finally, we performed yeast two-hybrid experiments to examine if CEH-30 interacts with UNC-37 and if the eh1/FIL motif of CEH-30 mediates the interaction of CEH-30 with UNC-37. In these assays, we fused either CEH-30 or UNC-37 to the GAL4 activation domain (AD) or DNA-binding domain (DB) and tested if the fusion proteins interact in yeast to induce the synthesis of the HIS3 and URA3 enzymes as well as the synthesis of β-D-galactosidase (lacZ). We found that yeast strains expressing both GAL4-AD-CEH-30 and GAL4-DB-UNC-37 or expressing both GAL4-DB-CEH-30 and GAL4-AD-UNC-37 grew on selective plates lacking histidine and containing 10 mM 3-amino-1,2,4-triazole (3AT, an inhibitor of the HIS3 enzyme), failed to grow on selective plates with 0.2% 5-fluoroorotic acid (5FOA, which is converted to toxic 5-fluorouracil by URA3), and had significantly elevated β-galactosidase activity (Fig. 5B). In contrast, yeast strains expressing GAL4-AD-CEH-30(F42E)/GALDB-UNC-37 or GAL4-DB-CEH-30(F42E)/GAL-AD-UNC-37 showed the opposite phenotype in each assay (Fig. 5B). These biochemical data indicate that CEH-30 and UNC-37 can physically interact through the eh1/FIL motif in CEH-30 and that CEH-30 inhibits CEM apoptosis in males by recruiting the transcriptional repressor UNC-37 to the promoter of its target gene(s).
Discussion
In a genetic screen for factors that regulate the sexually dimorphic apoptosis of CEM neurons, we identified ceh-30, a BarH homeodomain-encoding gene, as a key inhibitor of CEM cell death. Loss-of-function mutations in ceh-30 lead to ectopic CEM death in males. Genetic epistasis analysis indicates that ceh-30 acts downstream from the terminal sex determination factor tra-1, upstream of, or in parallel to, the cell death initiator egl-1, and probably upstream of the caspase ced-3 to control the sex-specific death of the CEM neurons (Table 2). Therefore ceh-30 defines a critical checkpoint that interprets sex specification signals and translates them into an appropriate life versus death response, probably by regulating the expression of a key cell death gene.
BarH homeodomain proteins were first identified in Drosophila due to the “Bar” mutant phenotype and later in several vertebrate species including fish, amphibians, and mammals. They are expressed primarily in the central nervous system, where they are implicated in regulating neuronal cell fates, migration, and survival (Reig et al. 2007). In C. elegans, ceh-30 appears to play a similar role in controlling the cell fate of the chemosensory CEM neurons, promoting the survival of CEMs in males. Interestingly, deletion of the mouse Barhl1 gene, which encodes a BarH homeodomain protein, leads to progressive degeneration of cochlear hair cells and loss of many neurons from the zonal layer of the superior colliculus due to increased apoptosis in these regions (Li et al. 2002; Li and Xiang 2006). These observations suggest that BarH homeodomain proteins may play a conserved role in promoting the survival of sensory neurons.
Analysis of two ceh-30 alleles (sm130 and tm272) reveals a cis-regulatory element within the second intron of the ceh-30 gene that is critical for the survival of CEMs in males. This cis-element, In2-POU, resembles a conserved binding site for POU-type homeodomain proteins. Alteration or removal of this In2-POU site by the sm130 mutation or the tm272 deletion causes ectopic death of CEMs in males, underscoring its importance for CEM survival. Interestingly, from the same CEM mutant screen, we identified a loss-of-function mutation (sm117) in the unc-86 gene that causes complete absence of CEM neurons. Given that unc-86 encodes a POU-type homeodomain protein and is expressed in CEM neurons (Finney et al. 1988; Finney and Ruvkun 1990), UNC-86 is an excellent candidate for an In2-POU-binding factor. Indeed, UNC-86 binds In2-POU in vitro, and the sm130 mutation reduces the binding of UNC-86 to the In2-POU site in a manner consistent with the weak reduction of CEM survival observed in sm130 males. Moreover, the mouse ortholog of UNC-86, Brn3c, also functions genetically upstream of Barhl1, a CEH-30 homolog, to specify terminal differentiation and survival of mechanosensory hair cells (Li et al. 2002). These findings suggest that UNC-86/Brn3c and CEH-30/Barhl1 may define an evolutionarily conserved pathway controlling the survival of sensory neurons.
How then does ceh-30 regulate sex-specific death of CEMs? Interestingly, a conserved TRA-1A-binding site (In2-TRA) is found 55 bp downstream from In2-POU and is altered in several gain-of-function ceh-30 mutants where CEMs improperly survive in hermaphrodites (H.T. Schwartz and H.R. Horvitz, pers. comm.), suggesting that In2-TRA may control sex-specific death of CEMs in response to the activity of TRA-1A. Indeed, we found that TRA-1A binds loosely to In2-TRA in vitro by itself and the binding of TRA-1A to this site is significantly enhanced in the presence of UNC-86, suggesting that UNC-86 interacts with TRA-1 to stabilize TRA-1A binding to intron 2. TRA-1A acts globally in the soma of the hermaphrodite to specify sexual fate. In the developing hermaphrodite, high levels of TRA-1A expression in CEMs presumably would result in binding of TRA-1A to intron 2 of the ceh-30 gene, which would interfere with or block the transcriptional activation of ceh-30 by UNC-86 that also binds intron 2. On the other hand, low or no expression of TRA-1A in developing males would allow UNC-86 to activate ceh-30 expression without interference from TRA-1A and thus promote CEM survival (Fig. 6A). It is worth noting that TRA-1A has also been shown to regulate the sexually dimorphic apoptosis of HSNs by binding to a cis-element in the egl-1 gene and negatively regulating the expression of egl-1 in hermaphrodite HSNs (Conradt and Horvitz 1999). Therefore, TRA-1A controls appropriate sexually dimorphic apoptosis of HSNs and CEMs by repressing the expression of two downstream target genes with contrasting apoptotic functions: the death-initiating egl-1 gene in HSNs and the death-inhibiting ceh-30 gene in CEMs (Fig. 6B).
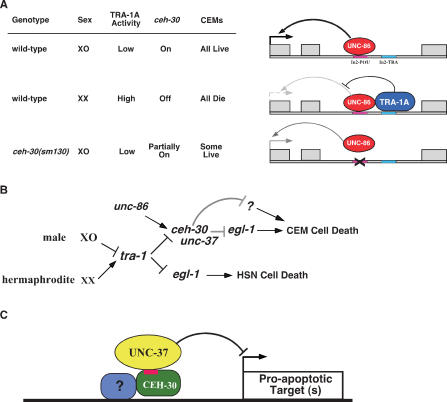
Figure 6.
Transcriptional regulation of sex-specific CEM cell death. (A) UNC-86, a transcription activator, and TRA-1A, a transcription repressor, interact at intron 2 of the ceh-30 gene to regulate the expression of ceh-30, leading to the proper cell death specification of CEMs. XO indicates males, and XX indicates hermaphrodites. (B) Genetic pathways regulating sexually dimorphic cell deaths in C. elegans. ceh-30 is the key gene that interprets the sex determination signal transduced by tra-1 and the survival signal specified by unc-86 to turn off or on programmed cell death in CEMs along with unc-37, probably through suppressing the expression of egl-1 and/or another proapoptotic gene. In HSNs, TRA-1A directly inhibits the expression of egl-1. (C) Formation of a CEH-30/UNC-37 repressosome at the promoter of a proapoptotic target gene. A CEM-specific transcription factor that binds to the promoter may recruit the UNC-37/Groucho transcription repressor through CEH-30 via its eh1/FIL motif (in red).
Given the importance of homeodomain proteins in cell fate determination, it is very surprising that the ceh-30 deletion allele (tm2157), which removes the CEH-30 homeodomain, has only a weak effect on the survival of male CEMs. This finding suggests that the homeodomain of CEH-30 is largely dispensable for its CEM death protective function and that the N terminus of CEH-30 is sufficient to confer most of its death protective activity. The N terminus of CEH-30 contains a conserved eh1/FIL motif that has been shown to mediate protein–protein interaction and recruitment of the general transcriptional repressor Groucho (Smith and Jaynes 1996; Choi et al. 1999; Jimenez et al. 1999; Bae et al. 2003). Indeed, this eh1/FIL motif in CEH-30 mediates interaction of CEH-30 with the C. elgans Groucho ortholog UNC-37 (Fig. 5B), and a mutation (F42E) that alters the conserved and critical Phe residue in this eh1/FIL motif disrupts the binding of CEH-30 to UNC-37 and the death protective function of CEH-30 in male CEMs. Furthermore, a mutation (e262) that reduces the activity of UNC-37 not only can cause ectopic male CEM death on its own but also can synergize with the ceh-30(tm2157) mutation to cause ectopic death of most male CEMs, suggesting that CEH-30 and UNC-37 act together to inhibit CEM cell death in males. It is interesting that both ceh-30 and unc-37 are widely expressed in C. elegans embryos (Fig. 4A; Winnier et al. 1999), yet ceh-30 specifically represses cell death only in male CEMs. This observation and the finding that the CEH-30 homeodomain is largely dispensable for its death inhibitory activity in CEMs suggest that CEH-30 may act to bridge the repressosome formation of UNC-37/Groucho with a CEM-specific transcription factor that directs repressosome binding to the promoter of a critical proapoptotic target gene and thus represses CEM cell death in males (Fig. 6C). In total, our study reveals a delicate transcriptional regulatory cascade that responds to appropriate sexual signals to repress or activate apoptosis in a sex-specific and cell-type-specific manner, thereby generating appropriate male-specific neurons (Fig. 6B).
Materials and methods
Strains
C. elegans strains were maintained at 20°C, unless otherwise noted. The N2 (Bristol) strain was the standard wild-type strain. For single-nucleotide polymorphism (SNP) mapping, the Hawaiian strain CB4856 was used. The alleles that were used in this study are LGI: ceh-6(mg60), unc-37(e262), bli-4(e937), dpy-5(e61), unc-13(e450), and hT2(I;III); LGII: smIs23; LGIII: tra-1(e1099), unc-86(sm117, n845, e1416, e1507), and dpy-18(e364); LGIV: smIs26, him-8(e1489), and ced-3(n717, n2433); LGV: egl-1(n1084n3082), him-5(e1490), and unc-76(e911); and LGX: ceh-30(sm130, tm272, tm2157), lon-2(e678), unc-2(e55), dpy-3(e27), ceh-18(mg57), and smIs54.
EMS mutagenesis
EMS mutagenesis was carried out as described previously (Brenner 1974). Briefly, mixed-stage worms were exposed to 47 mM EMS for 4 h with agitation. The F1 progeny of mutagenized animals were cloned out, and their F2 progeny were screened for loss of CEMs in males and improper survival in hermaphrodites.
Mapping of sm130
sm130 was mapped to LGX by crossing smIs23; him-5(e1490); sm130 males into smIs23; him-5(e1490); lon-2(e678) hermaphrodites, picking non-Lon cross progeny, and scoring the male progeny of Lon F2 animals for the CEM defect. Further three-factor mapping was done by crossing smIs23; him-5(e1490); sm130 males into unc-2(e55) lon-2(e678) hermaphrodites and scoring Lon non-Unc or Unc non-Lon F2 progeny. The location of sm130 was refined by consecutive SNP mapping using several SNP markers: snp_F53B3, snp_F52E4, snp_K09C4, snp_F11D5, and snp_C02F12 (Wicks et al. 2001). To obtain X-chromosome recombinants between sm130 animals and the SNP-rich Hawaiian strain (CB4856), smIs26 males carrying the X chromosome derived from CB4856 were crossed into smIs26; him-5(e1490); dpy-3(e27) sm130 lon-2(e678) hermaphrodites. Lon non-Dpy homozygous recombinants were analyzed for the presence of sm130 by scoring for CEM loss in males, and SNPs were genotyped by PCR amplification and subsequent restriction enzyme digestions. This SNP mapping strategy placed sm130 between two SNPs on Cosmids F53B3 and F52E4.
Scoring of CEMs and HSNs
The presence of CEMs was scored in L4 larvae and adults using the integrated Ppkd-2gfp reporter, smIs23, or smIs26. CEMs can also be scored in L4 larvae based on their cell morphology using Nomarski optics. The percentage of surviving CEMs was calculated by dividing the total number of CEMs observed by the maximum possible number of CEMs [(the number of CEMs observed)/(4 × the number of animals scored)]. The presence of HSNs was scored in L4 larvae using smIs26, which also carries Ptph-1gfp that directs GFP expression in HSNs (Sze et al. 2000) or by Nomarski optics in L1 or L4 larvae based on HSN cell morphology.
Molecular biology
C33D12 was digested with NheI, and the resulting 9496-bp ceh-30-containing fragment was cloned into the pBluescript SK(−) vector (Stratagene) via its XbaI site. This construct was then digested with SacI to remove a 1786-bp fragment, leaving a 7710-bp C33D12 NheI–SacI fragment. Pceh-30ceh-30∷gfp was constructed by PCR-amplifying the ceh-30 promoter and ORF from the first C33D12 9.5-kb construct using a ceh-30-specific primer 5′-GCTCTAGATTCTGAGTTGCTGGAAACATCC-3′ (contains an XbaI site) and a primer from the vector (T7: 5′-AATACGACTCACTATAG-3′). The resulting 5.8-kb PCR product was subcloned into pPD95.77 via its PstI and XbaI sites. Full-length ceh-30 cDNA was generated by RT–PCR using primers 5′-GCGATATCCTATTCTGAGTTGCTGGAAACAT CC-3′ (contains an EcoRV site) and 5′-GCGCTAGCATGTCA CTTCTCGACCCTCGGC-3′ (contains an NheI site). The amplified cDNA fragment was subcloned into pSL1190 (Amersham Biosciences) via its NheI and EcoRV sites. Pceh-30ceh-30(Δintron2)∷gfp was generated by PCR amplification of the ceh-30 cDNA using primers 5′-GCGCTAGCATGTCACTTC TCGACCCTCGGC-3′ and 5′-GCTCTAGATTCTGAGTTGC TGGAAACATCC-3′ (contains an XbaI site) followed by digestion with KpnI (within exon 2) and XbaI. The resulting KpnI–XbaI fragment was subcloned into Pceh-30ceh-30∷gfp that had been partially digested with KpnI and XbaI. Pceh-30ceh-30(ΔIn2-POU)∷gfp, Pceh-30ceh-30(1–92)∷gfp, and Pceh-30ceh-30(F42E)∷gfp constructs were generated using Stratagene’s QuikChange Site-Directed Mutagenesis Kit with the following primers: 5′-AATT TGTGCCTCTTTTGAAATGTATCCACAATCTGGAAAACC TTTCAAAACGT-3′ and 5′-GACGTTTTGAAAGGTTTTCCA GATTGTGGATACATTTCAAAAGAGGCACAAAT-3′ for making Pceh-30ceh-30(ΔIn2-POU)∷gfp, 5′-CCAGGCAGTTGCTAA TAATCTAGAAAGGCAAGAACC-3′ and 5′-GGTTCTTGCC TTTCTAGATTATTAGCAACTGCCTGG-3′ for making Pceh-30 ceh-30(1–92)∷gfp, and 5′-CAAAATTTCGTCTTCTTCTTCA GAGCGGATATCTGACATTCTCGAGCAATCC-3′ and 5′-G GGATTGCTCGAGAATGTCAGATATCCGCTCTGAAGAA GAAGACGAAATTTT-3′ for Pceh-30ceh-30(F42E)∷gfp.
Transgenic animals
Germline transformation was performed as described previously (Mello et al. 1991). Cosmid DNA, rescuing constructs, and GFP fusion reporters (20 ng/μL each) were injected into animals with the appropriate genetic background using Psur-5RFP as a coinjection marker, which directs expression of red fluorescent protein (RFP) in every cell of C. elegans larvae (Gu et al. 1998).
Expression and purification of proteins
Full-length UNC-86 was purified as described previously (Xue et al. 1993). Full-length tra-1 cDNA was subcloned into the pET-30b vector (Novagen) via its KpnI and NotI sites. BL21(DE3) Lys S bacterial cells were transformed with the appropriate expression vector, cultured to an OD600 of 0.4–0.5, induced to express the protein with 1 mM IPTG for 1 h at room temperature. Cells were then lysed by sonication in buffer A (10% glycerol, 20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 5 mM imidazole, 0.1% NP-40, and 1 mM PMSF). The soluble fraction was incubated with Ni2+-NTA-agarose beads for 4 h at 4°C. The beads were washed with 50 vol of wash buffer (buffer A with 60 mM imidazole and no PMSF). Bound proteins were eluted with buffer A containing 250 mM imidazole but no PMSF.
Gel mobility shift assays
For gel mobility shift analysis, oligonucleotides or PCR fragments were labeled with 32P-ATP using T4 polynucleotide kinase. Purified proteins (25 ng of GST∷UNC-86 and/or 5 ng of TRA-1∷His6) were preincubated with 0.05 mg/mL poly(dG-dC) in a buffer containing 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, and 10 mM Tris-HCl (pH 7.5) for 10 min at room temperature. Labeled DNA was then added and incubated for another 20 min at room temperature. Samples were resolved on 4% or 8% native polyacrylamide gels at 4°C. Gels were dried and exposed to PhosphorImaging screens overnight. Quantification of gel shift bands was done with the ImageQuant software package (GE Healthcare). Antibody supershift experiments were performed as described previously (Xue et al. 1993).
Yeast two hybrid
Yeast two-hybrid assays were performed using the ProQuest Two-Hybrid System with Gateway Technology (Invitrogen). GAL4 fusion proteins were generated using the following PCR primers for unc-37 and ceh-30, respectively: 5′-GGGGACAA GTTTGTACAAAAAAGCAGGCTTCATGAAGGCATCGTA TCTGG-3′, 5′-GGGGACCACTTTGTACAAGAAAGCTGGG TCTTAATATTCAACTGCATAGA-3′, 5′-GGGGACAAGTTT GTCAAAAAAGCAGGCTTCATGTCACTTCTCGACCCTC GG-3′, and 5′-GGGGACCACTTTGTACAAGAAAGCTGGG TCCTATTCTGAGTTGCTGGAAACATCC-3′. Yeast growth was assayed on synthetic complete SC-Leu-Trp plates lacking histidine but containing 10 mM 3AT, or lacking uracil while containing 0.2% 5FOA. The X-gal staining and the liquid β-galactosidase assay using chlorophenolred-β-D-galactopyranoside (CPRG) as a substrate were performed as described in the ProQuest manual.
Acknowledgments
We thank Maureen M. Barr and Paul W. Sternberg for the Ppkd-2gfp construct, David M. Miller for unc-37 strains and reagents, Hillel Schwartz and H. Robert Horvitz for communicating unpublished data on ceh-30(gf) mutants and ceh-30 epistasis analysis, and other Xue laboratory members for discussions and comments on the manuscript. This work is supported by a Burroughs Wellcome Fund Career Award and NIH R01 grants (GM66262 and GM59083) to D.X., and a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.M.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1607807
References
- Aoki M.P., Aoki A., Maldonado C.A., Aoki A., Maldonado C.A., Maldonado C.A. Sexual dimorphism of apoptosis in lactotrophs induced by bromocryptine. Histochem. Cell Biol. 2001;116:215–222. doi: 10.1007/s004180100307. [DOI] [PubMed] [Google Scholar]
- Argenton F., Ramoz N., Charlet N., Bernardini S., Colombo L., Bortolussi M., Ramoz N., Charlet N., Bernardini S., Colombo L., Bortolussi M., Charlet N., Bernardini S., Colombo L., Bortolussi M., Bernardini S., Colombo L., Bortolussi M., Colombo L., Bortolussi M., Bortolussi M. Mechanisms of transcriptional activation of the promoter of the rainbow trout prolactin gene by GHF1/Pit1 and glucocorticoid. Biochem. Biophys. Res. Commun. 1996;224:57–66. doi: 10.1006/bbrc.1996.0984. [DOI] [PubMed] [Google Scholar]
- Bae Y.K., Shimizu T., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Shimizu T., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Yabe T., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Kim C.H., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Hirata T., Nojima H., Muraoka O., Hirano T., Hibi M., Nojima H., Muraoka O., Hirano T., Hibi M., Muraoka O., Hirano T., Hibi M., Hirano T., Hibi M., Hibi M. A homeobox gene, pnx, is involved in the formation of posterior neurons in zebrafish. Development. 2003;130:1853–1865. doi: 10.1242/dev.00418. [DOI] [PubMed] [Google Scholar]
- Barr M.M., Sternberg P.W., Sternberg P.W. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Blush J., Lei J., Ju W., Silbiger S., Pullman J., Neugarten J., Lei J., Ju W., Silbiger S., Pullman J., Neugarten J., Ju W., Silbiger S., Pullman J., Neugarten J., Silbiger S., Pullman J., Neugarten J., Pullman J., Neugarten J., Neugarten J. Estradiol reverses renal injury in Alb/TGF-β1 transgenic mice. Kidney Int. 2004;66:2148–2154. doi: 10.1111/j.1523-1755.2004.66005.x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin T.R., Ruvkun G., Ruvkun G. Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development. 2001;128:779–790. doi: 10.1242/dev.128.5.779. [DOI] [PubMed] [Google Scholar]
- Casteels K.M., Gysemans C.A., Waer M., Bouillon R., Laureys J.M., Depovere J., Mathieu C., Gysemans C.A., Waer M., Bouillon R., Laureys J.M., Depovere J., Mathieu C., Waer M., Bouillon R., Laureys J.M., Depovere J., Mathieu C., Bouillon R., Laureys J.M., Depovere J., Mathieu C., Laureys J.M., Depovere J., Mathieu C., Depovere J., Mathieu C., Mathieu C. Sex difference in resistance to dexamethasone-induced apoptosis in NOD mice: Treatment with 1,25(OH)2D3 restores defect. Diabetes. 1998;47:1033–1037. doi: 10.2337/diabetes.47.7.1033. [DOI] [PubMed] [Google Scholar]
- Chin-Sang I.D., Spence A.M., Spence A.M. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes & Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- Choi C.Y., Kim Y.H., Kwon H.J., Kim Y., Kim Y.H., Kwon H.J., Kim Y., Kwon H.J., Kim Y., Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 1999;274:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R., Horvitz H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R., Horvitz H.R. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Doniach T., Hodgkin J., Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Dunbar M.E., Dann P.R., Robinson G.W., Hennighausen L., Zhang J.P., Wysolmerski J.J., Dann P.R., Robinson G.W., Hennighausen L., Zhang J.P., Wysolmerski J.J., Robinson G.W., Hennighausen L., Zhang J.P., Wysolmerski J.J., Hennighausen L., Zhang J.P., Wysolmerski J.J., Zhang J.P., Wysolmerski J.J., Wysolmerski J.J. Parathyroid hormone-related protein signaling is necessary for sexual dimorphism during embryonic mammary development. Development. 1999;126:3485–3493. doi: 10.1242/dev.126.16.3485. [DOI] [PubMed] [Google Scholar]
- Ellis H.M., Horvitz H.R., Horvitz H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H.R., Ruvkun G., Horvitz H.R., Horvitz H.R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- Forger N.G., Rosen G.J., Waters E.M., Jacob D., Simerly R.B., de Vries G.J., Rosen G.J., Waters E.M., Jacob D., Simerly R.B., de Vries G.J., Waters E.M., Jacob D., Simerly R.B., de Vries G.J., Jacob D., Simerly R.B., de Vries G.J., Simerly R.B., de Vries G.J., de Vries G.J. Deletion of Bax eliminates sex differences in the mouse forebrain. Proc. Natl. Acad. Sci. 2004;101:13666–13671. doi: 10.1073/pnas.0404644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein P. The synaptonemal complexes of Caenorhabditis elegans: Pachytene karyotype analysis of hermaphrodites from the recessive him-5 and him-7 mutants. J. Cell Sci. 1986;82:119–127. doi: 10.1242/jcs.82.1.119. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Hird S., Plasterk R.H., Andachi Y., Kohara Y., Wang B., Finney M., Ruvkun G., Hird S., Plasterk R.H., Andachi Y., Kohara Y., Wang B., Finney M., Ruvkun G., Plasterk R.H., Andachi Y., Kohara Y., Wang B., Finney M., Ruvkun G., Andachi Y., Kohara Y., Wang B., Finney M., Ruvkun G., Kohara Y., Wang B., Finney M., Ruvkun G., Wang B., Finney M., Ruvkun G., Finney M., Ruvkun G., Ruvkun G. Targeted mutations in the Caenorhabditis elegans POU homeo box gene ceh-18 cause defects in oocyte cell cycle arrest, gonad migration, and epidermal differentiation. Genes & Dev. 1994;8:1935–1948. doi: 10.1101/gad.8.16.1935. [DOI] [PubMed] [Google Scholar]
- Gu T., Orita S., Han M., Orita S., Han M., Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Scott M.P., Scott M.P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hengartner M.O., Ellis R.E., Horvitz H.R., Ellis R.E., Horvitz H.R., Horvitz H.R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Sexual dimorphism and sex determination in the nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. [DOI] [PubMed] [Google Scholar]
- Hodgkin J.A., Brenner S., Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- Jennings B.H., Pickles L.M., Wainwright S.M., Roe S.M., Pearl L.H., Ish-Horowicz D., Pickles L.M., Wainwright S.M., Roe S.M., Pearl L.H., Ish-Horowicz D., Wainwright S.M., Roe S.M., Pearl L.H., Ish-Horowicz D., Roe S.M., Pearl L.H., Ish-Horowicz D., Pearl L.H., Ish-Horowicz D., Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Jimenez G., Verrijzer C.P., Ish-Horowicz D., Verrijzer C.P., Ish-Horowicz D., Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol. Cell. Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kimble J., Edgar L., Hirsh D., Edgar L., Hirsh D., Hirsh D. Specification of male development in Caenorhabditis elegans: The fem genes. Dev. Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- Kuwabara P.E., Okkema P.G., Kimble J., Okkema P.G., Kimble J., Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Xiang M., Xiang M. Barhl1 is required for maintenance of a large population of neurons in the zonal layer of the superior colliculus. Dev. Dyn. 2006;235:2260–2265. doi: 10.1002/dvdy.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Price S.M., Cahill H., Ryugo D.K., Shen M.M., Xiang M., Price S.M., Cahill H., Ryugo D.K., Shen M.M., Xiang M., Cahill H., Ryugo D.K., Shen M.M., Xiang M., Ryugo D.K., Shen M.M., Xiang M., Shen M.M., Xiang M., Xiang M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development. 2002;129:3523–3532. doi: 10.1242/dev.129.14.3523. [DOI] [PubMed] [Google Scholar]
- Mehra A., Gaudet J., Heck L., Kuwabara P.E., Spence A.M., Gaudet J., Heck L., Kuwabara P.E., Spence A.M., Heck L., Kuwabara P.E., Spence A.M., Kuwabara P.E., Spence A.M., Spence A.M. Negative regulation of male development in Caenorhabditis elegans by a protein–protein interaction between TRA-2A and FEM-3. Genes & Dev. 1999;13:1453–1463. doi: 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V., Kramer J.M., Stinchcomb D., Ambros V., Stinchcomb D., Ambros V., Ambros V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.J. Sex determination and X chromosome dosage compensation. In: Riddle D.L., et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 209–240. [PubMed] [Google Scholar]
- Parrish J., Metters H., Chen L., Xue D., Metters H., Chen L., Xue D., Chen L., Xue D., Xue D. Demonstration of the in vivo interaction of key cell death regulators by structure-based design of second-site suppressors. Proc. Natl. Acad. Sci. 2000;97:11916–11921. doi: 10.1073/pnas.210391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M.D., Li W., Trent C., Robertson B., Fire A., Hageman J.M., Wood W.B., Li W., Trent C., Robertson B., Fire A., Hageman J.M., Wood W.B., Trent C., Robertson B., Fire A., Hageman J.M., Wood W.B., Robertson B., Fire A., Hageman J.M., Wood W.B., Fire A., Hageman J.M., Wood W.B., Hageman J.M., Wood W.B., Wood W.B. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes & Dev. 1993;7:216–228. doi: 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- Pflugrad A., Meir J.Y., Barnes T.M., Miller D.M., Meir J.Y., Barnes T.M., Miller D.M., Barnes T.M., Miller D.M., Miller D.M. The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Price J.M., Donahoe P.K., Ito Y., Hendren W.H., Donahoe P.K., Ito Y., Hendren W.H., Ito Y., Hendren W.H., Hendren W.H. Programmed cell death in the Mullerian duct induced by Mullerian inhibiting substance. Am. J. Anat. 1977;149:353–375. doi: 10.1002/aja.1001490304. [DOI] [PubMed] [Google Scholar]
- Reig G., Cabrejos M.E., Concha M.L., Cabrejos M.E., Concha M.L., Concha M.L. Functions of BarH transcription factors during embryonic development. Dev. Biol. 2007;302:367–375. doi: 10.1016/j.ydbio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Rhee J.M., Gruber C.A., Brodie T.B., Trieu M., Turner E.E., Gruber C.A., Brodie T.B., Trieu M., Turner E.E., Brodie T.B., Trieu M., Turner E.E., Trieu M., Turner E.E., Turner E.E. Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem. 1998;273:34196–34205. doi: 10.1074/jbc.273.51.34196. [DOI] [PubMed] [Google Scholar]
- Roberts L.M., Shen J., Ingraham H.A., Shen J., Ingraham H.A., Ingraham H.A. New solutions to an ancient riddle: Defining the differences between Adam and Eve. Am. J. Hum. Genet. 1999;65:933–942. doi: 10.1086/302601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S., Bargmann C.I., Bargmann C.I. Control of neuronal subtype identity by the C. elegans ARID protein CFI-1. Genes & Dev. 2002;16:972–983. doi: 10.1101/gad.976002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.T., Jaynes J.B., Jaynes J.B. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N., Schierenberg E., White J.G., Thomson J.N., White J.G., Thomson J.N., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sze J.Y., Victor M., Loer C., Shi Y., Ruvkun G., Victor M., Loer C., Shi Y., Ruvkun G., Loer C., Shi Y., Ruvkun G., Shi Y., Ruvkun G., Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Gish W.R., Waterston R.H., Plasterk R.H., Waterston R.H., Plasterk R.H., Plasterk R.H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Winnier A.R., Meir J.Y., Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., Meir J.Y., Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., Ross J.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., Tavernarakis N., Driscoll M., Ishihara T., Katsura I., Miller D.M., Driscoll M., Ishihara T., Katsura I., Miller D.M., Ishihara T., Katsura I., Miller D.M., Katsura I., Miller D.M., Miller D.M. UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes & Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Finney M., Ruvkun G., Chalfie M., Finney M., Ruvkun G., Chalfie M., Ruvkun G., Chalfie M., Chalfie M. Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. EMBO J. 1992;11:4969–4979. doi: 10.1002/j.1460-2075.1992.tb05604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Tu Y., Chalfie M., Tu Y., Chalfie M., Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- Yan N., Chai J., Lee E.S., Gu L., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., Chai J., Lee E.S., Gu L., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., Lee E.S., Gu L., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., Gu L., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., He J., Wu J.W., Kokel D., Li H., Hao Q., Wu J.W., Kokel D., Li H., Hao Q., Kokel D., Li H., Hao Q., Li H., Hao Q., Hao Q., et al. Structure of the CED-4–CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yin Y., Lin C., Ma L., Lin C., Ma L., Ma L. MSX2 promotes vaginal epithelial differentiation and wolffian duct regression and dampens the vaginal response to diethylstilbestrol. Mol. Endocrinol. 2006;20:1535–1546. doi: 10.1210/me.2005-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Hodgkin J., Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: A gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Hodgkin J., Hodgkin J. Zinc fingers in sex determination: Only one of the two C. elegans TRA-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993;21:3691–3698. doi: 10.1093/nar/21.16.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]