Abstract
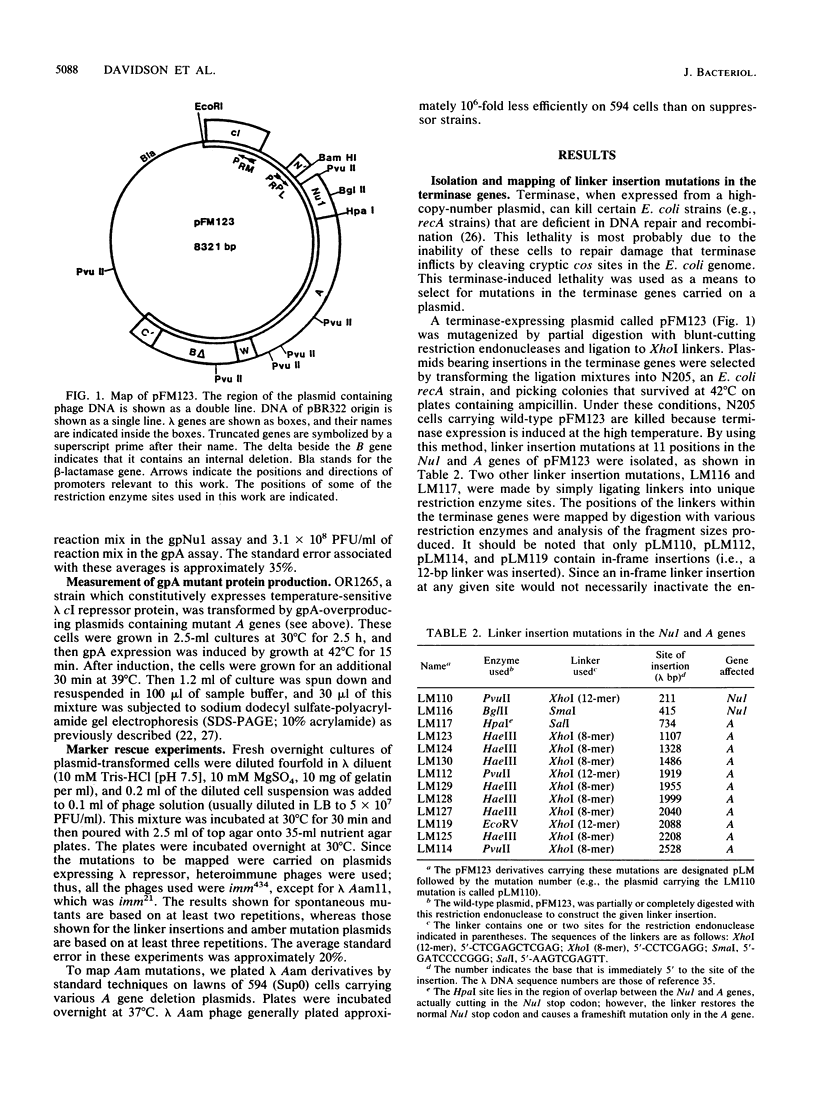
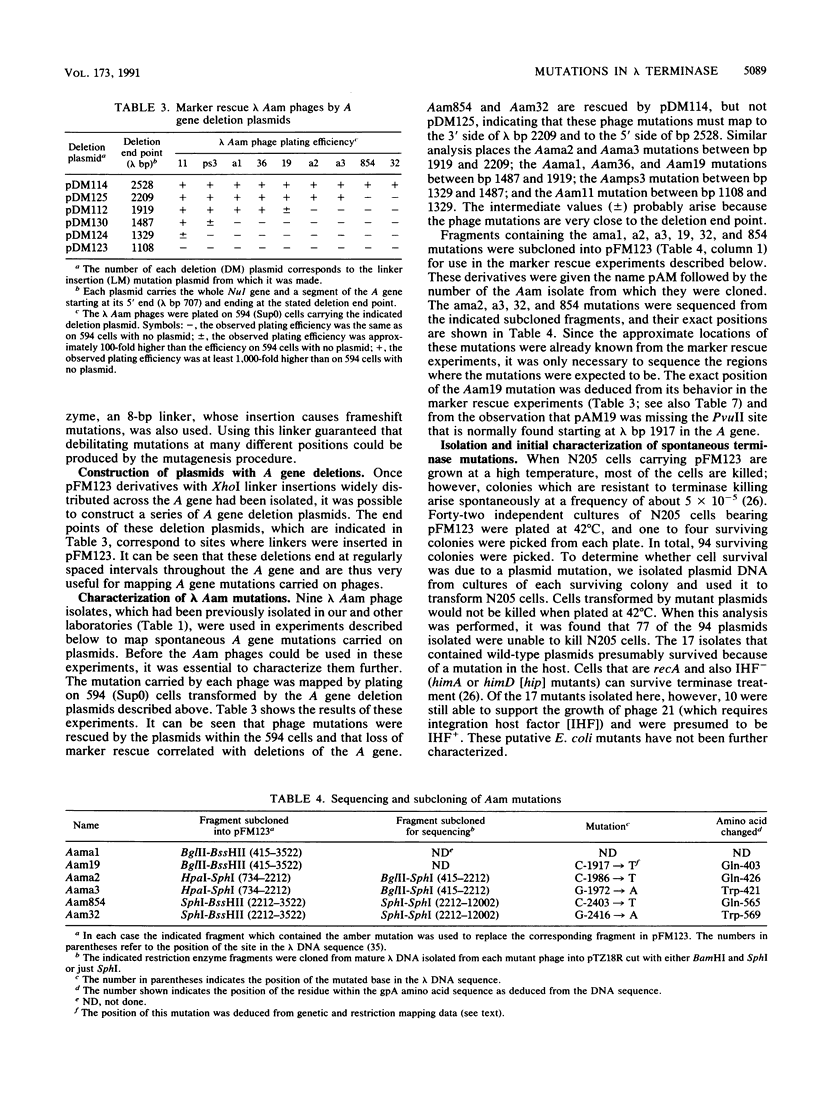
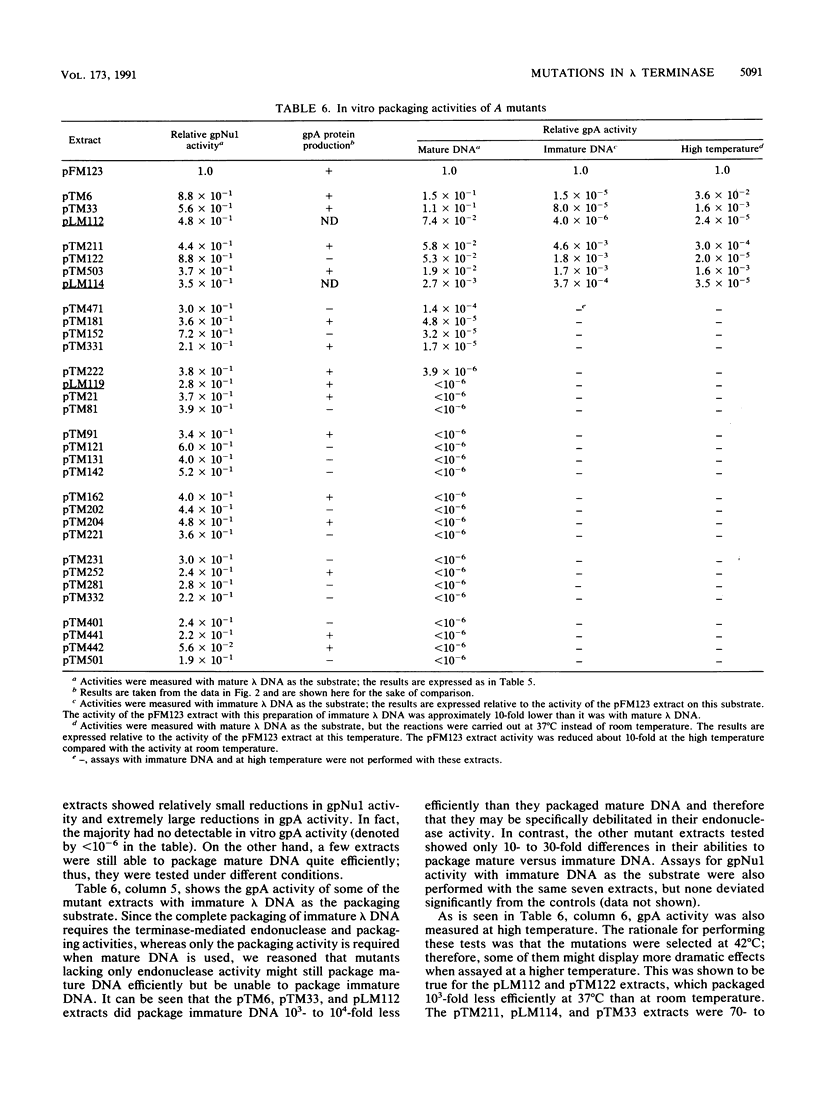
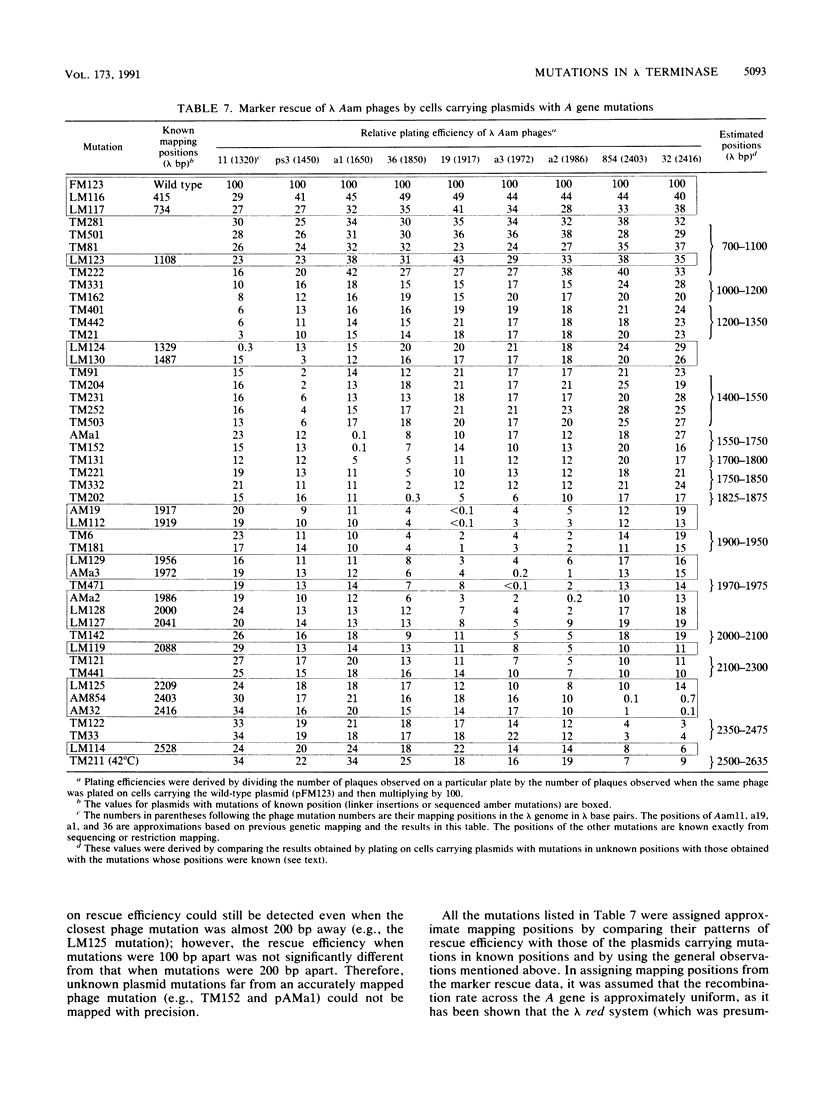
The terminase enzyme of bacteriophage lambda is a hetero-oligomeric protein which catalyzes the site-specific endonucleolytic cleavage of lambda DNA and its packaging into phage proheads; it is composed of the products of the lambda Nul and A genes. We have developed a simple method to select mutations in the terminase genes carried on a high-copy-number plasmid, based on the ability of wild-type terminase to kill recA strains of Escherichia coli. Sixty-three different spontaneous mutations and 13 linker insertion mutations were isolated by this method and analyzed. Extracts of cells transformed by mutant plasmids displayed variable degrees of reduction in the activity of one or both terminase subunits as assayed by in vitro lambda DNA packaging. A method of genetically mapping plasmid-borne mutations in the A gene by measuring their ability to rescue various lambda Aam phages showed that the A mutations were fairly evenly distributed across the gene. Mutant A genes were also subcloned into overproducing plasmid constructs, and it was determined that more than half of them directed the synthesis of normal amounts of full-length A protein. Three of the A gene mutants displayed dramatically reduced in vitro packaging activity only when immature (uncut) lambda DNA was used as the substrate; therefore, these mutations may lie in the endonuclease domain of terminase. Interestingly, the putative endonuclease mutations mapped in two distinct locations in the A gene separated by a least 400 bp.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker A., Gold M. Isolation of the bacteriophage lambda A-gene protein. Proc Natl Acad Sci U S A. 1975 Feb;72(2):581–585. doi: 10.1073/pnas.72.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Gold M. Prediction of an ATP reactive center in the small subunit, gpNu1, of the phage lambda terminase enzyme. J Mol Biol. 1988 Jan 5;199(1):219–222. doi: 10.1016/0022-2836(88)90391-9. [DOI] [PubMed] [Google Scholar]
- Becker A., Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990 Jun;172(6):2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Murialdo H., Gold M. Studies on an in vitro system for the packaging and maturation of phage lambda DNA. Virology. 1977 May 1;78(1):277–290. doi: 10.1016/0042-6822(77)90099-x. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Bittner M., Vapnek D. Versatile cloning vectors derived from the runaway-replication plasmid pKN402. Gene. 1981 Dec;15(4):319–329. doi: 10.1016/0378-1119(81)90175-x. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Chow S., Daub E., Murialdo H. The overproduction of DNA terminase of coliphage lambda. Gene. 1987;60(2-3):277–289. doi: 10.1016/0378-1119(87)90236-8. [DOI] [PubMed] [Google Scholar]
- Frackman S., Siegele D. A., Feiss M. A functional domain of bacteriophage lambda terminase for prohead binding. J Mol Biol. 1984 Dec 5;180(2):283–300. doi: 10.1016/s0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- Frackman S., Siegele D. A., Feiss M. The terminase of bacteriophage lambda. Functional domains for cosB binding and multimer assembly. J Mol Biol. 1985 May 25;183(2):225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- Gold M., Becker A. The bacteriophage lambda terminase. Partial purification and preliminary characterization of properties. J Biol Chem. 1983 Dec 10;258(23):14619–14625. [PubMed] [Google Scholar]
- Gold M., Parris W. A bacterial protein requirement for the bacteriophage lambda terminase reaction. Nucleic Acids Res. 1986 Dec 22;14(24):9797–9809. doi: 10.1093/nar/14.24.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Peterson C., Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi 29. J Mol Biol. 1987 Sep 20;197(2):229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Higgins R. R., Lucko H. J., Becker A. Mechanism of cos DNA cleavage by bacteriophage lambda terminase: multiple roles of ATP. Cell. 1988 Sep 9;54(6):765–775. doi: 10.1016/s0092-8674(88)91021-5. [DOI] [PubMed] [Google Scholar]
- Jara L., Murialdo H. Isolation of nonsense mutants in the morphogenetic region of the bacteriophage lambda chromosome. Virology. 1975 Mar;64(1):264–268. doi: 10.1016/0042-6822(75)90097-5. [DOI] [PubMed] [Google Scholar]
- Kimura M., Fujisawa H. Dissection of functional domains of the packaging protein of bacteriophage T3 by site-directed mutagenesis. Virology. 1991 Feb;180(2):709–715. doi: 10.1016/0042-6822(91)90084-o. [DOI] [PubMed] [Google Scholar]
- Kypr J., Mrázek J. Lambda phage protein Nu 1 contains the conserved DNA binding fold of repressors. J Mol Biol. 1986 Sep 5;191(1):139–140. doi: 10.1016/0022-2836(86)90430-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McClure S. C., MacHattie L., Gold M. A sedimentation analysis of DNA found in Escherichia coli infected with phage lambda mutants. Virology. 1973 Jul;54(1):1–18. doi: 10.1016/0042-6822(73)90109-8. [DOI] [PubMed] [Google Scholar]
- Mendelson I., Gottesman M., Oppenheim A. B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991 Mar;173(5):1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Feiss M. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol Gen Genet. 1988 Apr;212(1):157–165. doi: 10.1007/BF00322459. [DOI] [PubMed] [Google Scholar]
- Murialdo H., Davidson A., Chow S., Gold M. The control of lambda DNA terminase synthesis. Nucleic Acids Res. 1987 Jan 12;15(1):119–140. doi: 10.1093/nar/15.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H., Fife W. L., Becher A., Feiss M., Yochem J. Bacteriophage lambda DNA maturation. The functional relationships among the products of genes Nul, A and FI. J Mol Biol. 1981 Jan 15;145(2):375–404. doi: 10.1016/0022-2836(81)90211-4. [DOI] [PubMed] [Google Scholar]
- Murialdo H. Lethal effect of lambda DNA terminase in recombination deficient Escherichia coli. Mol Gen Genet. 1988 Jul;213(1):42–49. doi: 10.1007/BF00333396. [DOI] [PubMed] [Google Scholar]
- Murialdo H., Siminovitch L. The morphogenesis of bacteriophage lambda. IV. Identification of gene products and control of the expression of the morphogenetic information. Virology. 1972 Jun;48(3):785–823. doi: 10.1016/0042-6822(72)90162-6. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Genetics of the left arm of the chromosome of bacteriophage lambda. Genetics. 1968 Jul;59(3):311–325. doi: 10.1093/genetics/59.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parris W., Davidson A., Keeler C. L., Jr, Gold M. The Nu1 subunit of bacteriophage lambda terminase. J Biol Chem. 1988 Jun 15;263(17):8413–8419. [PubMed] [Google Scholar]
- Parsell D. A., Sauer R. T. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989 May 5;264(13):7590–7595. [PubMed] [Google Scholar]
- Reyes O., Gottesman M., Adhya S. Formation of lambda lysogens by IS2 recombination: gal operon--lambda pR promoter fusions. Virology. 1979 Apr 30;94(2):400–408. doi: 10.1016/0042-6822(79)90470-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Shinder G., Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J Virol. 1988 Feb;62(2):387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Weisberg R. Packaging of coliphage lambda DNA. I. The role of the cohesive end site and the gene A protein. J Mol Biol. 1977 Dec 15;117(3):717–731. doi: 10.1016/0022-2836(77)90066-3. [DOI] [PubMed] [Google Scholar]
- Weigle J. Assembly of phage lambda in vitro. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. F., Christiansen S., Feiss M. Domains for protein-protein interactions at the N and C termini of the large subunit of bacteriophage lambda terminase. Genetics. 1988 Jul;119(3):477–484. doi: 10.1093/genetics/119.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Ito J. Nonsense codons and polarity in the tryptophan operon. J Mol Biol. 1966 Nov 14;21(2):313–334. doi: 10.1016/0022-2836(66)90102-1. [DOI] [PubMed] [Google Scholar]



