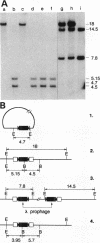
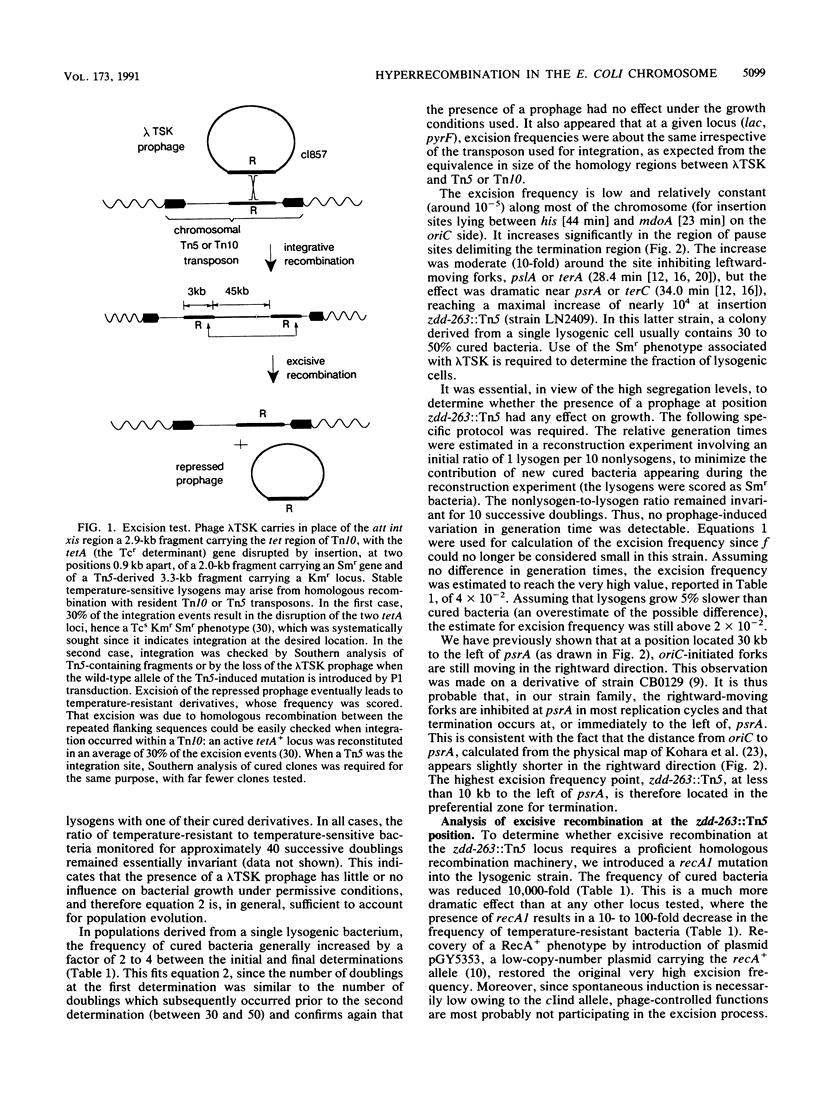
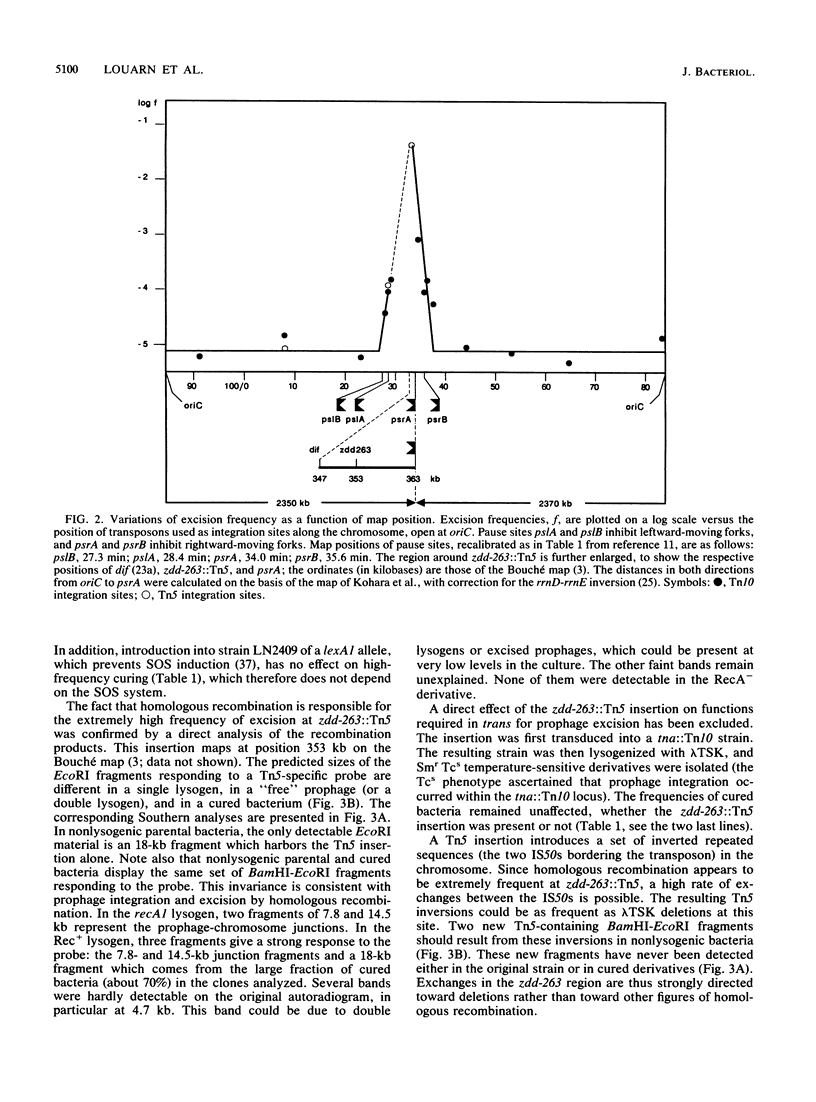
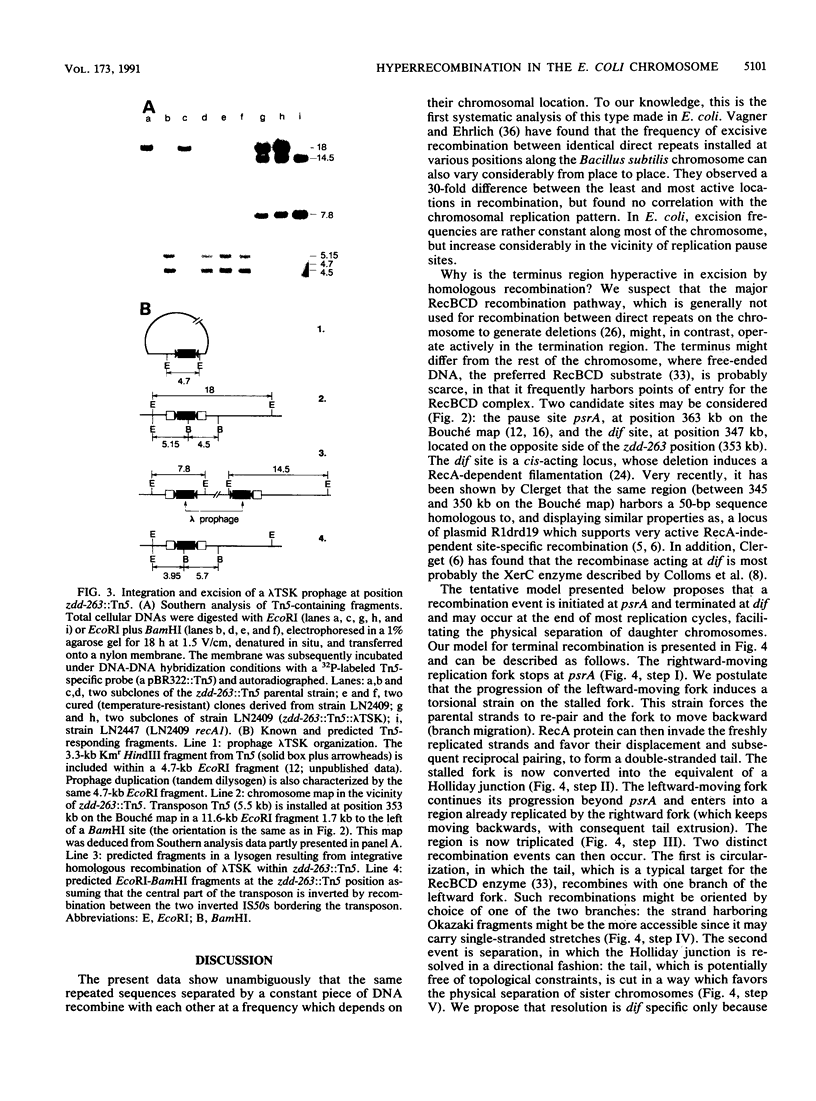
Abstract
The frequency of excisive homologous recombination has been measured at various positions along the Escherichia coli chromosome. The reporter system makes use of a lambda cI857 prophage integrated by homologous recombination within Tn5 or Tn10 transposons already installed at known positions in the E. coli chromosome. The excision frequency per cell and per generation was determined by monitoring the evolution of the relative number of temperature-resistant (cured) bacteria is a function of the age of the cultures. Excisions, due to RecA-dependent homologous exchanges, appeared to occur more frequently in the preferential termination zone for chromosome replication. The highest frequency of excision observed is compatible with a recombination event at each replication cycle in this region. On the basis of these data, we propose a model involving homologous recombination in the final steps of bacterial chromosome replication and separation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché J. P. Physical map of a 470 x 10(3) base-pair region flanking the terminus of DNA replication in the Escherichia coli K12 genome. J Mol Biol. 1982 Jan 5;154(1):1–20. doi: 10.1016/0022-2836(82)90413-2. [DOI] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Clerget M. A 140 base-pair DNA segment from the kanamycin resistance region of plasmid R1 acts as an origin of replication and promotes site-specific recombination. J Mol Biol. 1984 Sep 5;178(1):35–46. doi: 10.1016/0022-2836(84)90229-8. [DOI] [PubMed] [Google Scholar]
- Cohen A., Clark A. J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986 Jul;167(1):327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms S. D., Sykora P., Szatmari G., Sherratt D. J. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990 Dec;172(12):6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutreix M., Moreau P. L., Bailone A., Galibert F., Battista J. R., Walker G. C., Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989 May;171(5):2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François V., Conter A., Louarn J. M. Properties of new Escherichia coli Hfr strains constructed by integration of pSC101-derived conjugative plasmids. J Bacteriol. 1990 Mar;172(3):1436–1440. doi: 10.1128/jb.172.3.1436-1440.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François V., Louarn J., Louarn J. M. The terminus of the Escherichia coli chromosome is flanked by several polar replication pause sites. Mol Microbiol. 1989 Aug;3(8):995–1002. doi: 10.1111/j.1365-2958.1989.tb00250.x. [DOI] [PubMed] [Google Scholar]
- François V., Louarn J., Patte J., Louaran J. M. A system for in vivo selection of genomic rearrangements with predetermined endpoints in Escherichia coli using modified Tn10 transposons. Gene. 1987;56(1):99–108. doi: 10.1016/0378-1119(87)90162-4. [DOI] [PubMed] [Google Scholar]
- Henson J. M., Kuempel P. L. Deletion of the terminus region (340 kilobase pairs of DNA) from the chromosome of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3766–3770. doi: 10.1073/pnas.82.11.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M., Akiyama M., Horiuchi T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988 Nov 4;55(3):467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- Hill T. M., Henson J. M., Kuempel P. L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1754–1758. doi: 10.1073/pnas.84.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Kopp B. J., Kuempel P. L. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J Bacteriol. 1988 Feb;170(2):662–668. doi: 10.1128/jb.170.2.662-668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Marians K. J. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. M., Pelletier A. J., Tecklenburg M. L., Kuempel P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988 Nov 4;55(3):459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Khatri G. S., MacAllister T., Sista P. R., Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989 Nov 17;59(4):667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lee E. H., Kornberg A., Hidaka M., Kobayashi T., Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan M. J., Roth J. R. Role of recBC function in formation of chromosomal rearrangements: a two-step model for recombination. Genetics. 1989 Mar;121(3):433–443. doi: 10.1093/genetics/121.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo J. E., François V., Louarn J. M. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9391–9395. doi: 10.1073/pnas.85.24.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles B., Paoletti C. Control of UV induction of recA protein. Proc Natl Acad Sci U S A. 1983 Jan;80(1):65–69. doi: 10.1073/pnas.80.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Vagner V., Ehrlich S. D. Efficiency of homologous DNA recombination varies along the Bacillus subtilis chromosome. J Bacteriol. 1988 Sep;170(9):3978–3982. doi: 10.1128/jb.170.9.3978-3982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- de Massy B., Béjar S., Louarn J., Louarn J. M., Bouché J. P. Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1759–1763. doi: 10.1073/pnas.84.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]