Summary
Terminase enzymes are common to double-stranded DNA (dsDNA) viruses and are responsible for packaging viral DNA into the confines of an empty capsid shell. In bacteriophage lambda the catalytic terminase subunit is gpA, which is responsible for maturation of the genome end prior to packaging and subsequent translocation of the matured DNA into the capsid. DNA packaging requires an ATPase catalytic site situated in the N-terminus of the protein. A second ATPase catalytic site associated with the DNA maturation activities of the protein has been proposed; however, direct demonstration of this putative second site is lacking. Here we describe biochemical studies that define protease-resistant peptides of gpA and expression of these putative domains in E. coli. Biochemical characterization of gpA-ΔN179, a construct in which the N-terminal 179 residues of gpA have been deleted, indicates that this protein encompasses the DNA maturation domain of gpA. The construct is folded, soluble and possesses an ATP-dependent nuclease activity. Moreover, the construct binds and hydrolyzes ATP despite the fact that the DNA packaging ATPase site in the N-terminus of gpA has been deleted. Mutation of lysine 497, which alters the conserved lysine in a predicted Walker A “P-loop” sequence, does not affect ATP binding but severely impairs ATP hydrolysis. Further, this mutation abrogates the ATP-dependent nuclease activity of the protein. These studies provide direct evidence for the elusive nucleotide-binding site in gpA that is directly associated with the DNA maturation activity of the protein. The implications of these results with respect to the two roles of the terminase holoenzyme – DNA maturation and DNA packaging – are discussed.
Keywords: DNA packaging, motor protein, virus assembly, terminase
Introduction
Terminase enzymes are common to many dsDNA viruses of both prokaryotic and eukaryotic origin1; 2; 3; 4. These enzymes are responsible for the ATP-dependent insertion of viral DNA into an empty procapsid, a process called DNA packaging. The preferred packaging substrate is often a linear DNA concatemer composed of multiple genomes linked in a head-to-tail fashion (immature DNA1)3; 4; 5; 6. The terminase enzymes excise an individual genome from the concatemer and concomitantly translocate the “matured” DNA into the capsid. All of the characterized terminase enzymes are hetero-oligomers composed of a large subunit, which provides the packaging activities of the enzyme, and a small subunit that is responsible for specific recognition of viral DNA2; 3; 5; 6. In the case of bacteriophage lambda, the functional terminase holoenzyme is composed of gpA and gpNu1 subunits in a heterotrimeric gpA1●gpNu12 protomer complex7; 8. The larger gpA subunit (73.3 kDa, 641 amino acids) possesses all of the catalytic activities required for DNA maturation and packaging6; 9. These include site specific endonuclease (cos-cleavage) and strand separation (helicase) activities required to excise an individual genome from the concatemer (DNA maturation activity), DNA translocase (packaging) activity and ATPase activity9; 10; 11; 12; 13; 14; 15; 16. Genetic and biochemical studies have suggested that the DNA packaging (translocase) and the DNA maturation (nuclease/helicase) catalytic activities reside in discrete N-terminal and C-terminal domains, respectively, of the gpA subunit17; 18; 19; 20; 21. While the packaging ATPase catalytic site has been clearly identified, direct demonstration of a discrete ATPase activity associated with DNA maturation has been elusive.
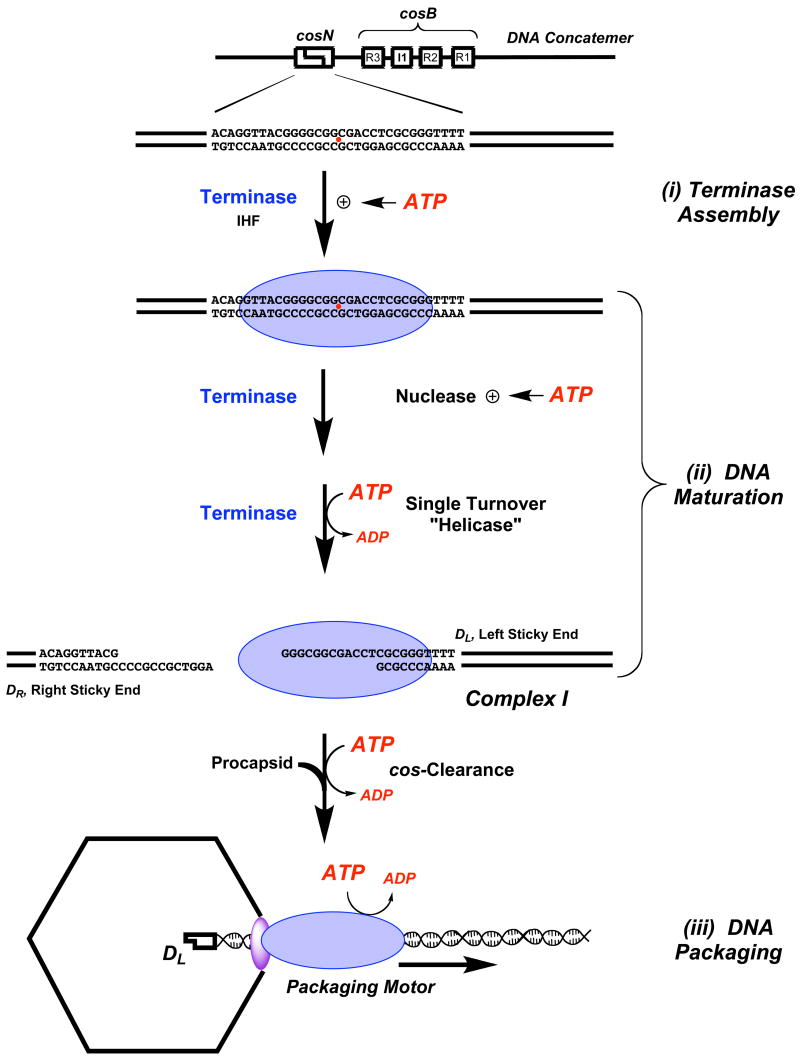
The cohesive end sequence, or cos, is the junction between individual genomes in concatemeric lambda DNA and is the site where terminase assembles to initiate the packaging process6; 9. Assembly of the packaging machinery is mediated by the smaller gpNu1 subunit, which specifically recognizes the three R-elements found within the cosB subsite (Figure 1)9; 22; 23. This presumably positions a dimer of gpA subunits symmetrically disposed at the cosN half-sites, which introduce site-specific nicks into the duplex twelve bases apart (the cos cleavage reaction)9; 10; 13; 16. The so-called “helicase” activity of gpA then separates the nicked, annealed strands ejecting the DR end from the enzyme; duplex unwinding is non-processive, however, and does not proceed beyond the 12 bases of single-stranded DNA (“sticky” end) generated by the cos cleavage reaction (see Figure 1)9; 12; 24; 25. Mechanistically, this may be viewed as a single catalytic turnover helicase reaction that affords complex I, an incredibly stable nucleoprotein complex composed of terminase tightly bound to the mature single-stranded left end of lambda DNA (DL “sticky” end, Figure 1)25.
Figure 1. DNA Maturation and DNA Packaging by Lambda Terminase.
Genetic organization of the cos site in a lambda concatemer is shown at top. Terminase holoenzyme (a gpA1●gpNu12 heterotrimer) is represented as a blue oval. (i) Terminase Assembly at cos. Cooperative binding of gpA (at cosN) and gpNu1 (at cosB) is modulated by ATP. (ii) DNA Maturaton. The rate and fidelity of the cos-cleavage reaction is stimulated by ATP and ATP hydrolysis fuels the strand separation (helicase) reaction to yield complex I. (iii) DNA Packaging. The gpA subunits in complex I bind to the portal ring situated in a unique vertex of the icosahedral procapsid (purple oval), which triggers cos-clearance and activation of the DNA translocase activity of gpA. The packaging motor translocates DNA into the capsid interior powered by ATP hydrolysis.
Complex I next binds to the portal, a doughnut-shaped ring situated at a unique vertex of an empty procapsid; this interaction triggers two critical events. (i) Terminase switches from a cos-specific single-stranded DNA binding protein to a mobile complex that binds tightly, but non-specifically to duplex DNA, and (ii) the packaging activity of the enzyme is activated, which translocates DNA into the capsid14; 19; 26; 27; we presume that the nuclease activity of the enzyme is simultaneously inactivated to prevent duplex nicking during packaging. The transition from the site-specifically bound complex I to the mobile packaging motor is referred to as cos-clearance, in analogy to promoter clearance by RNA polymerase enzymes28. The activated packaging motor translocates DNA into the capsid until the next cos sequence (the end of the genome) is encountered. At this point, DNA translocation stops and the maturation functions are activated such that the terminase gpA subunit again nicks the duplex at cosN and separates the strands releasing the DNA-filled capsid and re-generating complex I. Finishing proteins and a tail are added to the capsid, which affords an infectious virus; the re-generated complex I binds to another empty procapsid to initiate a second round of processive packaging9.
ATP plays multiple roles in the packaging process (see Figure 1); ATP modulates the rate and fidelity of the cos-cleavage reaction29; 30, ATP hydrolysis drives the strand separation reaction12; 24 and DNA packaging is fueled by the hydrolysis of ATP12; 26; 27; 31. It is further likely that ATP and/or ATP hydrolysis regulates terminase assembly at cos to initiate packaging and subsequently triggers cos-clearance upon procapsid binding (vide infra). It has long been appreciated that the functional terminase holoenzyme possesses ATPase activity and sequence analysis suggested the presence of Walker A phosphate-binding loops, or “P-loops”, in each of the terminase subunits32; 33. This motif is composed of a number of glycine residues and a conserved lysine that is essential to NTP binding and/or hydrolysis34; 35. The presence of P-loop consensus sequences centered at Lys35 and Lys497 of the gpNu1 and gpA subunits, respectively, suggested that both of the terminase subunits might possess ATPase activity; this was confirmed by mutagenesis and kinetic experiments, which identified a low-affinity ATPase catalytic site in gpNu1 (Km~ 1.2 mM minus DNA; Km~ 500 μM plus DNA) and a high-affinity site in gpA (Km~ 5 μM)11; 36. The role of the ATPase catalytic site in gpNu1 is unclear, but it is likely involved in terminase assembly at the cos-site and regulation of terminase release from cos to initiate DNA packaging (i.e., cos clearance) (Figure 1)9; 37. It had been presumed that the high-affinity ATPase site in gpA is primarily involved in fueling DNA translocation during packaging9; 18. Unexpectedly, however, mutation of the conserved lysine in the predicted Walker A P-loop of gpA (K497) does not significantly affect ATP hydrolysis or DNA packaging by terminase holoenzyme17. Rather, the gpA-K497 mutations result in a defect in DNA maturation steps including cos-cleavage and strand separation by the holoenzyme17. We note that the effect of these mutations in the isolated gpA subunit has not been investigated.
The above experiments opened to question the nature of the putative Walker A P-loop motif at Lys497 (WA2, Figure 2A), which led to biochemical studies designed to physically locate the ATP binding site in gpA. Photoaffinity labeling experiments clearly showed that gpA is covalently modified by 8-azido ATP (AzATP), a photo reactive ATP analog, both in context of the holoenzyme18; 38 and in the isolated protein37. Unexpectedly, however, residues surrounding Lys497 are not labeled. Rather, Tyr46 and Lys84 in the N-terminus of the protein are covalently modified18, localizing them proximate to an ATP binding site. Close inspection of the primary sequence of gpA identified a second, albeit atypical Walker A motif in this region of the protein (WA1, Figure 2A)39. Mutational analysis confirmed that alteration of either Tyr46 or Lys84 significantly attenuated AzATP photolabeling of gpA and abrogated ATP hydrolysis by the subunit. Moreover, the mutant viruses had severe defects in virion assembly18. In sum, the data conclusively demonstrate that the biochemically characterized high-affinity ATPase site resides in the N-terminus of gpA, proximate Tyr46/Lys84. The data further indicate that this catalytic site is directly linked to the DNA packaging activity of protein. We therefore refer to this physically and kinetically identified site as the packaging ATPase site.
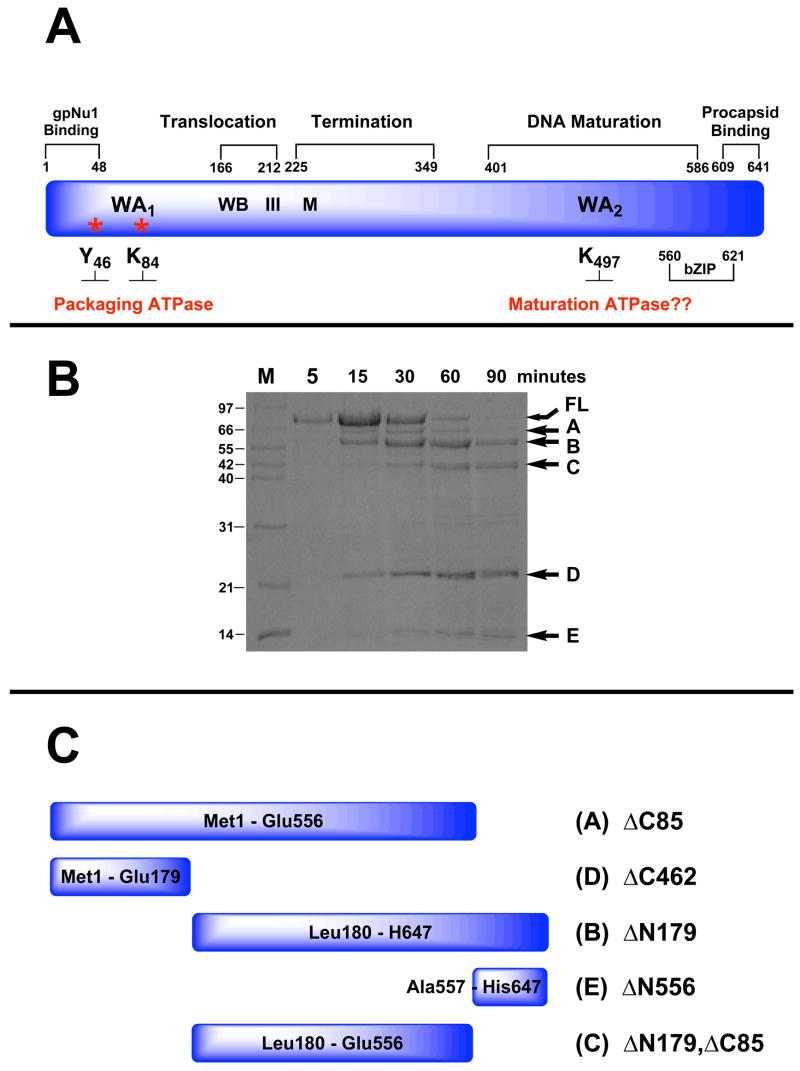
Figure 2.
Panel A. Genetic Organization of gpA. The full-length protein (641 residues) is depicted in blue and functional domains identified by genetic studies are indicated above. Walker A and B sequences (WA1, WB) identified by primary sequence analysis and associated with the packaging ATPase site are indicated. Residues covalently modified with AzATP (Y46, K84) are indicated with asterisks. Helicase motif III and metal binding (M) residues identified by primary sequence analysis are also indicated. The putative Walker A sequence in the C-terminal DNA maturation ATPase site (WA2 identified by primary sequence analysis) is also indicated. Panel B. Limited Proteolysis of gpA by Endo-Glu-C. The isolated gpA subunit was digested with Endo-Glu-C as described in Materials and Methods and the time course monitored by SDS-PAGE. Lane M, protein molecular weight standards with sizes indicated to the left of the gel. The undigested, full-length protein (FL) and protease resistant fragments (A–E) are indicated with arrows at right. Panel C. Cartoon depicting the deduced sequence of the gpA protease-resistant peptides.
In addition to its role in DNA packaging, ATP modulates the nuclease activity of gpA and ATP hydrolysis is required to drive the strand separation reaction (Figure 1)9; 24. Interestingly, however, the Tyr46 and Lys84 mutations that abrogate the ATPase and DNA packaging activities of gpA (vide supra) affect neither the nuclease nor the helicase activities of the mutant holoenzymes18. Based on the ensemble of genetic, mutagenesis and biochemical data, Feiss and co-workers proposed that the gpA subunit possesses a second “catalytically silent” ATPase site, distinct from the packaging-associated ATPase located in the N-terminus of the protein17; 21. According to the model, this site resides within the DNA maturation domain of gpA and is responsible for regulation of the cos-cleavage reaction and for powering the strand separation activity of the enzyme. Presumably, the second site is centered at Lys497 and is appropriately defined by the Walker A P-loop motif originally identified by primary sequence analysis (WA2, Figure 2A). We refer to this hypothetical site as the DNA maturation ATPase site. Direct evidence for a DNA maturation ATPase site in gpA has remained elusive, however.
Here we describe biochemical experiments that define protease-resistant fragments of gpA and expression of several of these constructs in E. coli. These studies have defined a DNA maturation domain of the protein that possesses nuclease activity. Importantly, we show that the nuclease activity of the construct is dependent on ATP, that the protein binds and hydrolyzes ATP and that mutation of K497, the conserved lysine in the putative P-loop motif, severely impairs ATP hydrolysis. These studies provide direct evidence for a second nucleotide-binding site in gpA that is directly associated with the DNA maturation activity of the protein. The implications of these results with respect to the mechanism of viral DNA maturation and packaging are discussed.
Results
Limited Proteolysis of the Lambda Terminase gpA Subunit
Genetic studies have suggested a domain organization for gpA and we used limited proteolysis techniques to probe for structural domains of the protein. Chymotrypsin, trypsin, proteinase K and Endo-Glu-C were each used to probe for protease-resistant fragments as described in Materials and Methods. The four enzymes were chosen to ensure that similar results are obtained using proteases with different substrate specificities. Indeed, qualitatively similar results were obtained with all four proteases (not shown) and a representative digestion time course obtained with Endo Glu-C is shown in Figure 2B. The data show that five prominent Endo Glu-C resistant peptides appear during the digestion time course, which are labeled A – E in the figure. Neither ATP nor DNA affects the proteolysis pattern or the digestion time course with either protease (data not shown). The identities of fragments A – E were determined by N-terminal sequence analysis, mass spectrometry and the predicted proteolysis pattern of the gpA subunit (Table S1, Supplementary Information); the deduced sequences of the peptides are shown in cartoon representation in Figure 2C.
Construction, Expression and Purification of gpA Deletion Constructs
Vectors that over express the five protease-resistant peptide fragments shown in Figure 2C were constructed as described in Materials and Methods. The deletion constructs were expressed in E. coli and supernatants from the crude cell lysates were examined for the presence of soluble protein. Deletion constructs that retained the N-terminus of gpA (i.e., gpA-ΔC85, gpA-ΔC462) were completely insoluble (Table S1, Supplementary Information). The proteins could be solubilized in 6 M guanidinium hydrochloride, but again precipitated upon dialysis of the denaturant from the buffer (data not shown). The gpA-ΔN179, ΔC85 construct, which lacks 179 and 85 residues from the N-terminus and C-terminus of the protein, respectively (Figure 2C), is partially soluble but aggregates at elevated protein concentrations (not shown). The small C-terminal fragment gpA-ΔN556 and the N-terminal deletion construct gpA-ΔN179 were both found in the soluble fraction of the crude cell lysate (Table S1, Supplementary Information).
In addition to the above proteins we also constructed a vector that expresses the gpA-ΔN179 deletion construct containing a Lys to Asp mutation at position 497 (numbering system for the full-length protein); this mutation alters the conserved lysine within the putative Walker A P-loop motif of gpA (WA2, Figure 2A). This protein expresses well in E. coli and is similarly found in the soluble fraction of the crude cell lysate (not shown). The gpA-ΔN179 and gpA-ΔN179,K497D proteins were each purified to greater than 95% purity as determined by SDS-PAGE and gel filtration chromatography (see Figure S1, Supplementary Information).
Biochemical Characterization of the gpA Deletion Constructs
We next examined the biochemical properties of the purified gpA-ΔN179 and gpA-ΔN179,K497D deletion constructs. In sum, the data indicate that both proteins are folded, soluble and predominantly monomeric in the concentration range of 1 μM to 15 μM (see Supplementary Information). This is in contrast to the isolated full-length gpA subunit (gpA-FL), which is a mixture of oligomeric species in solution that irreversibly aggregate upon concentration (D. Bain and C.E. Catalano, unpublished). Notwithstanding, both deletion constructs possess spectral properties essentially identical to that of the isolated full-length subunit (Figure S1 and Table S2, Supplementary Information). The data further indicate that the Lys497->Asp mutation does not significantly affect the physical properties of gpA-ΔN179.
Fluorescence Monitored Nucleotide Binding and ATP Hydrolysis by Full-Length gpA
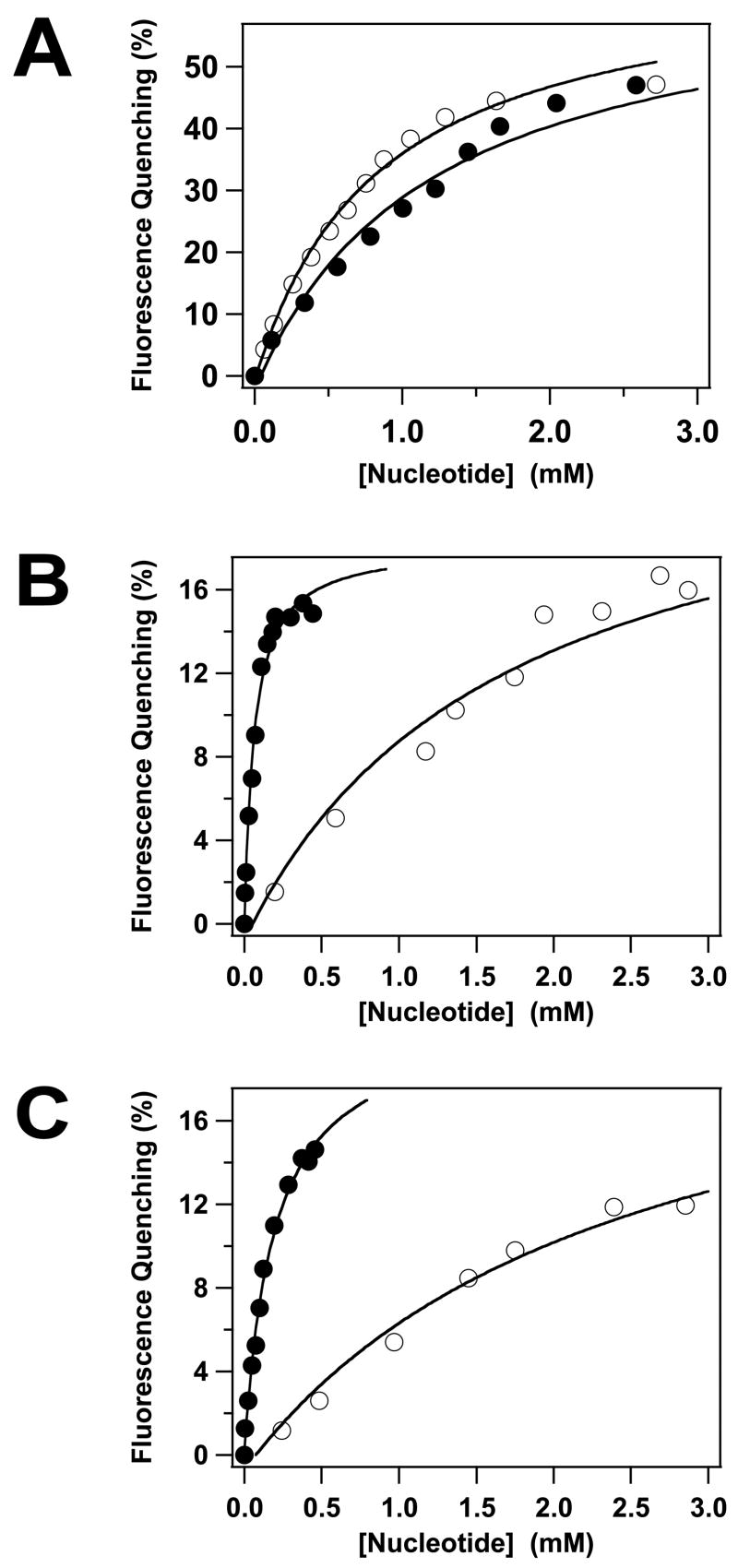
The gpA subunit possesses ATPase activity15; 40; 41; 42 and we used fluorescence spectroscopy as a probe for nucleotide binding to the isolated full-length subunit. ATP and ADP each induce a significant quenching of the intrinsic fluorescence of gpA-FL without a change in the λem,max (not shown); quenching by both nucleotides is concentration dependent and saturable (Figure 3A). Analysis of the data using a simple Langmuir binding model indicates that both nucleotides bind to the protein in the millimolar concentration range (Table 1). It is noteworthy that this is significantly greater than the Km~ 5 μM for ATP hydrolysis by this subunit - in the context of the holoenzyme11; 36. To address this discrepancy, we attempted to define the primary kinetic constants for ATP hydrolysis by gpA-FL. Unfortunately, these studies were frustrated by visible precipitation of the protein in the presence of ≥ 50 μM Mg2+●ATP (data not shown). Nonetheless, analysis of the data available ([ATP] < 50 μM) provides an estimation of the primary kinetic constants that are consistent with previously published values (kcat= 0.49 ± 0.1 min−1; Km= 70 ± 29 μM)40; 41; 42.
Figure 3.
Fluorescence-Monitored Nucleotide Binding to the gpA Deletion Constructs. ATP (○) or ADP (●) was incrementally added to gpA-FL (Panel A), gpA-ΔN179 (Panel B) or gpA-ΔN179,K497D (Panel C) and the intrinsic fluorescence of the proteins recorded as described in Materials and Methods. Each data point represents the average of at least three separate experiments (standard deviation 2%-3%, omitted for clarity). The solid lines represent the best fits of the data to a simple Langmuir binding model.
Table 1.
Nucleotide Binding and Hydrolysis by Full-Length gpA, gpA-ΔN179 and gpA-ΔN179,K497D
| Nucleotide | gpA-FL | gpA-ΔN179 | gpA-ΔN179,K497D |
|---|---|---|---|
| aATP | KD,app= 0.8 ± 0.1 mM | KD,app= 1.7 ± 0.6 mM | KD,app= 2.5 ± 0.6 mM |
| aADP | KD,app= 1.2 ± 0.2 mM | KD,app= 63 ± 10 μM | KD,app= 192 ± 26 μM |
| AMP | bN.C. | bN.C. | bN.C. |
| Single-Turnover
ATPase Activity |
ck1= 0.706 ± 0.089 min−1
k2= 0.017 ± 0.025 min−1 |
dkobs= 0.086 ± 0.008 min−1 | dkobs= 0.0072 ± 0.0004 min−1 |
The aggregation of gpA-FL in the presence of Mg2+●ATP is of interest. Further investigation revealed that the protein similarly aggregates in the presence of Mg2+●ADP (not shown). In contrast, the solubility of gpA-FL was unaffected by Mg2+●GTP, Mg2+●ATP-γS or Mg2+●AMP-PNP. Moreover, the protein remains soluble in the presence of Mg2+ alone and in the presence of nucleotides (up to 5 mM) in the absence of Mg2+. These data suggest that Mg2+●ADP, but not Mg2+●ATP, drive a conformational change that results in self-association of the protein.
Fluorescence Monitored Nucleotide Binding by the gpA Deletion Constructs
The high-affinity DNA packaging ATPase site (WA1) has been deleted in the gpA-Δ179 construct, but the protein retains the putative Walker A P-loop motif centered at Lys497 (WA2, Figure 2A). It is unclear whether the latter residues are part of a bona fide ATP binding site, however, and we therefore examined nucleotide binding by the deletion construct. ATP and ADP each induce a significant quenching of the intrinsic fluorescence of gpA-ΔN179 without a change in the λem,max (not shown); quenching by both nucleotides is concentration dependent and saturable (Figure 3B). Analysis of the data using a simple Langmuir binding model indicates that ATP binds to gpA-ΔN179 in the millimolar concentration range, similar to the full-length protein (Table 1). In contrast, however, ADP binds to the deletion construct with significantly greater affinity (Table 1). Similar to the full-length protein, the gpA-ΔN179 deletion construct also exhibits Mg2+-nucleotide dependent aggregation properties (not shown).
Essentially identical results were obtained with the gpA-ΔN179,K497D construct, though the nucleotide binding affinity is slightly reduced relative to gpA-ΔN179 (Table 1). This indicates that mutation of the conserved lysine within the putative WA2 P-loop motif does not significantly impair nucleotide binding to the protein.
ATP Hydrolysis by the gpA Deletion Constructs
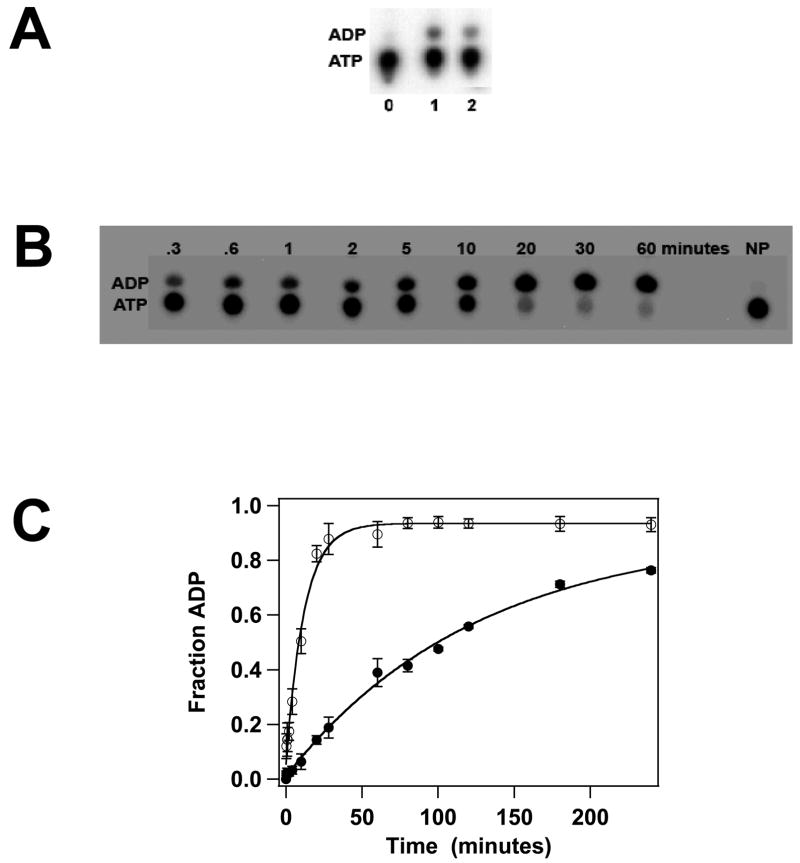
Previous biochemical studies have suggested that gpA contains a single ATPase catalytic site that is located in the N-terminus of the protein (WA1, Figure 2A)11; 36; 37. This region has been deleted in both gpA-ΔN179 and gpA-ΔN179,K497D, yet both proteins bind ATP and ADP. Therefore, we next examined the capacity of the constructs to hydrolyze ATP; both proteins possess a weak but detectable steady-state ATPase activity (Figure 4A) that is unaffected by DNA (not shown). The data suggest that the K497D mutation adversely affects ATPase activity, but the slow rate of ATP hydrolysis by both constructs makes steady-state kinetic analysis difficult and highly error-prone. To circumvent this problem, we examined ATP hydrolysis in a single-turnover kinetic experiment where the concentration of protein is in excess of ATP substrate. Under these conditions, a single catalytic turnover is observed in which the rate of ADP formation reflects ATP binding and hydrolysis steps only2. The data presented in Figures 4B and 4C clearly show that while both proteins possess ATPase activity, the K497D mutation severely compromises ATP hydrolysis, decreasing the observed rate by over an order of magnitude (Table 1).
Figure 4.
ATPase Activity of the gpA Deletion Constructs. Panel A. Steady-state ATP hydrolysis was conducted as described in Materials and Methods and a representative autoradiogram is shown. The migration positions of ATP and ADP on the TLC plate are indicated at left. Lane 0, no protein; lane 1, gpA-ΔN179; lane 2, gpA-ΔN179,K497D. Panel B. Single-turnover ATP hydrolysis was conducted as described in Materials and Methods and a representative autoradiogram for the gpA-ΔN179 deletion construct is shown. The migration positions of ATP and ADP on the TLC plate are indicated at left. NP no protein added to the reaction mixture. Panel C. Single-turnover ATPase data for gpA-ΔN179 (○) and gpA-ΔN179 (●) were quantified as described in Materials and method. Each data point represents the average of three separate experiments with the standard deviation indicated with error bars.
DNA Binding Activity of the gpA Deletion Constructs
The gpA-ΔN179 and gpA-ΔN179,K497D deletion constructs encompass the putative DNA maturation domain of full-length gpA and both are thus expected to interact with viral DNA. We therefore examined the DNA binding activity of these proteins using an electrophoretic mobility shift (EMS) assay. The results of these experiments are presented in Figure 5A and demonstrate that both constructs bind to cos-containing DNA yielding two diffuse retarded bands. Similar results are obtained with a DNA substrate of random sequence (not shown), suggesting that the constructs bind DNA in a sequence-independent manner. Analysis of the data indicates that both proteins bind DNA in the ~ 500 nM range (not shown). These results are essentially identical to those observed with the isolated full-length gpA subunit, which binds DNA in a non-specific manner to yield two retarded bands and in a similar concentration range25.
Figure 5.
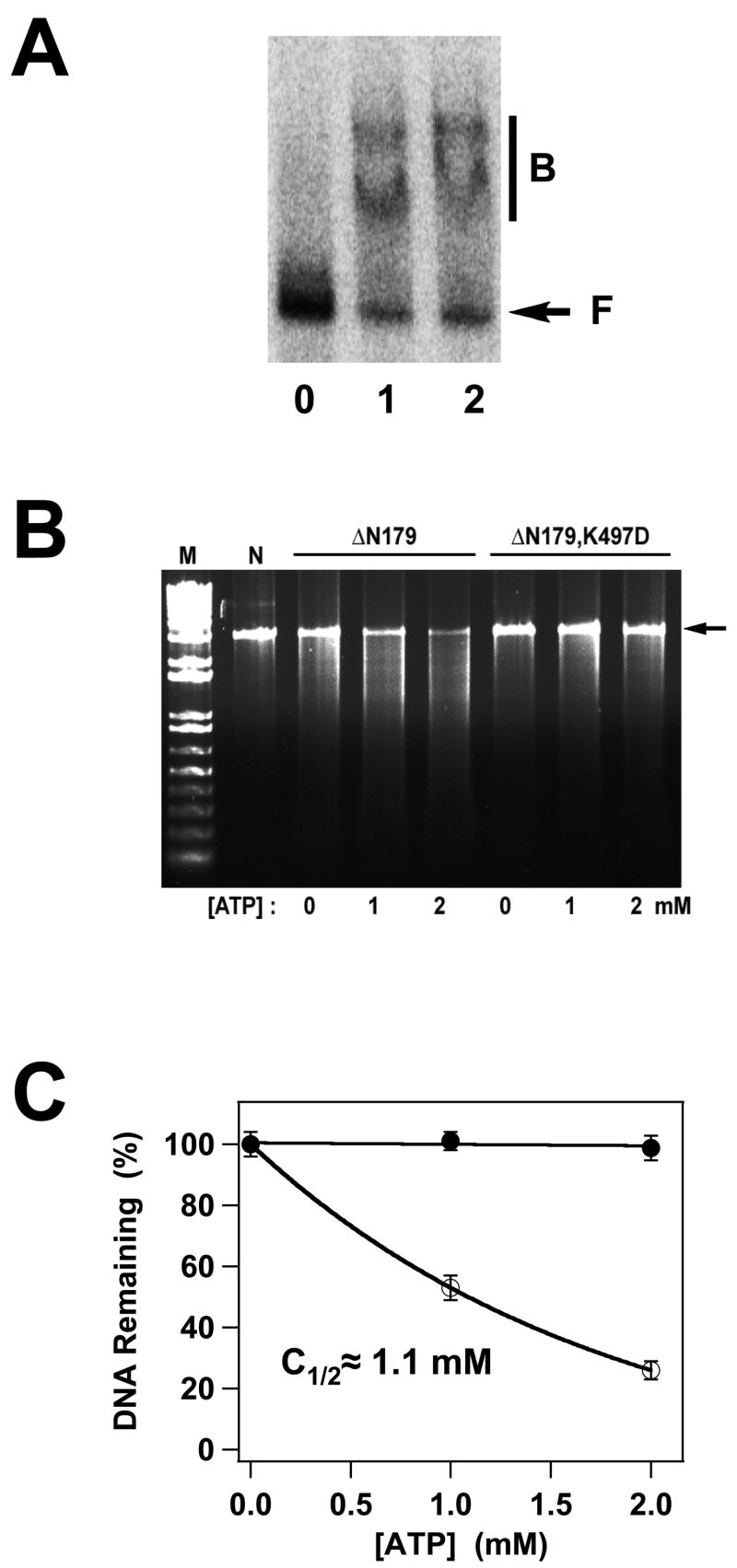
Panel A. DNA Binding by the gpA Deletion Constructs. The EMS experiment was conducted as described in Materials and Methods and a representative autoradiogram is shown. Lane 0, no protein; lane 1, gpA-ΔN179; lane 2, gpA-ΔN179,K497D. The migration of free DNA is indicated with an arrow (F), while bound DNA is indicated with a bar (B). Panel B. Nuclease Activity of the gpA Deletion Constructs. The nuclease assay was conducted as described in Materials and Methods and a representative gel is shown. Lanes M and N contain 1 kb DNA standards and the pAFP DNA substrate, respectively, in the absence of protein. The migration position of the DNA substrate is indicated with an arrow at right. GpA-ΔN179 or gpA-ΔN179,K497D proteins were included as indicated at the top of the figure and in the absence or presence of ATP as indicated at the bottom of the gel. The plasmid pAFP1 contains an intact cos site; specific cleavage at cos would afford 2 kb and 1 kb product bands, which are not visible in the gel. The nuclease activity of the protein is thus non-specific. Panel C. The data for gpA-ΔN179 (○) and gpA-ΔN179,K497D (●) presented in Panel B were quantified as described in Materials and Methods. Each data point represents the average of three separate experiments with the standard deviation indicated with error bars.
Nuclease/Helicase Activity of the gpA Deletion Constructs
The isolated full-length gpA subunit possesses little to no endonuclease activity, depending on the reaction conditions13; 40; 41; 42. The nuclease rate and specificity for the cos sequence is strongly activated by interaction with gpNu1 in the holoenzyme complex and is further modulated by ATP16; 29; 30; 37; 42. The gpA-ΔN179 deletion construct encompasses the putative nuclease domain of the protein and we next examined the nuclease activity of this construct. The protein possesses a non-specific nuclease activity that is dependent on ATP and that is stimulated in a concentration-dependent manner (Figure 5B). Half-maximal stimulation is achieved at an ATP concentration of ~ 1.1 mM (Figure 5C), which is similar to the KD,app for ATP binding determined by fluorescence quenching (Figure 3B, Table 1). Interestingly, while ADP binds tightly to the construct, it does not activate nuclease activity (data not shown).
Genetic and biochemical studies have shown that mutation of Lys497 in the WA2 P-loop motif of gpA abrogates the endonuclease activity of subunit - within the context of the holoenzyme17. Consistently, the gpA-ΔN179,K497D deletion construct possesses little, if any endonuclease activity, either in the absence or presence of ATP (Figures 5B and 5C).
The isolated full-length gpA subunit also possesses a weak strand separation activity that has been localized to the C-terminal portion of the protein (see Figure 2A)18; 21. The gpA deletion constructs retain this region and were therefore tested for strand separation activity as previously described24. An extremely weak but consistent activity was observed with the gpA-ΔN179 deletion construct that was dependent on ATP (not shown). Unfortunately, the non-specific nuclease activity of the protein resulted in significant degradation of the product bands, which precluded a detailed analysis of the reaction. In contrast, there was no evidence for strand separation activity with the gpA-ΔN179,K497D mutant construct (data not shown).
Discussion
Terminase enzymes are a central component of the DNA packaging motors in both prokaryotic and eukaryotic dsDNA viruses. In all known cases, the functional holoenzymes are hetero-oligomers composed of a large subunit that possess all of the catalytic activities required to package the genome and a small subunit that is required for specific recognition of viral DNA3. Despite intensive study, structural information of these enzymes remains limited, primarily due to insolubility of the purified proteins. We recently reported the high-resolution solution structure of the DNA binding domain of gpNu1, the small subunit of lambda terminase43. Identification of this domain was the culmination of limited proteolysis studies and extensive biochemical and biophysical characterization of soluble deletion constructs of the protein40; 41; 44.
Genetic and biochemical studies have suggested a domain organization for the lambda terminase gpA subunit that includes DNA packaging and DNA maturation domains located in the N-terminal and C-terminal thirds of the protein, respectively (see Figure 2A)9; 17; 18; 19; 20; 21. We utilized limited proteolysis methods to define structural domains in gpA that encompass these essential biological functions and the data presented here clearly demonstrate the presence of protease resistant fragments of the protein. Cloning and expression of several of these fragments yielded interesting observations, as follows. First, constructs retaining the N-terminal 179 residues of gpA are completely insoluble. Genetic studies have demonstrated that the N-terminal 48 residues of gpA interact with the gpNu1 subunit to form the terminase holoenzyme complex45. We propose that the N-terminal region of gpA is unstructured in the absence of gpNu1, which leads to significant aggregation of the isolated protein. Consistent with this hypothesis, the isolated full-length gpA subunit is polydisperse and aggregates when concentrated, while the N-terminal deletion constructs are soluble monomers. We note that the isolated gpNu1 subunit adopts a molten-globule-like structure in the absence of gpA, but attains significant structure and increased stability in its presence46. Thus, the subunit interacting domains in gpNu1 (C-terminus) and gpA (N-terminus) appear to require direct and specific interactions to assume a native protein fold. This concept is consistent with our recent demonstration that lambda terminase holoenzyme is a heterotrimer composed of one gpA molecule in stable association with a dimer of gpNu1 proteins (gpA1●gpNu12) and that the subunits dissociate only under denaturing conditions8.
The DNA Maturation Domain of gpA
Deletion of the N-terminal 179 residues of gpA affords a soluble construct that encompasses the entirety of the DNA maturation domain of gpA defined by genetic studies and additional residues that play an undefined role in termination of the packaging reaction (Figure 2A)19; 20. We note that termination requires duplex nicking and strand separation by gpA and these residues may, in fact, be part of an extended DNA maturation domain. The gpA-ΔN179 construct binds to DNA in a non-specific manner and with an affinity essentially identical to that of the full-length protein. The construct also possesses a non-specific nuclease activity that is dependent on ATP and we have thus defined a structural and functional DNA maturation domain of gpA.
Generalized Model for Terminase Domains
The results obtained here mirror those observed in the bacteriophage T4 system. The large subunit from T4 terminase (gp17) is similarly composed of an N-terminal DNA packaging domain and a C-terminal nuclease domain47, analogous to gpA. Limited-proteolysis of the T4 large terminase subunit cleaves the protein into two fragments – (i) an N-terminal half that possesses a high-affinity ATPase site and the DNA packaging activity of the protein, and (ii) a C-terminal half that possesses a non-specific nuclease activity47. These observations may reflect a functional conservation among the terminase holoenzymes. Indeed, sequence homology is observed among the terminase large subunits, particularly in the N-terminal regions, that is retained even in eukaryotic enzymes such as those in the herpesvirus groups39. These domains display Walker A, Walker B, adenine base-binding and catalytic carboxylate signatures (see Figure 2A)39. These “ATPase domains” exhibit features common to the ATPase domains of the DEAD box helicases, including the “motif III” sequence that couples ATP hydrolysis to DNA translocation48. The primary function of the terminase packaging domains is to translocate viral DNA into a pre-formed capsid, which requires that the motor complexes bind DNA tightly, but non-specifically. Sequence and structural homology between these domains and other non-specific DNA translocation machines is thus not surprising. Indeed, a recent crystal structure of the DNA packaging domain of bacteriophage T4 terminase reveals structural similarities with monomeric helicases 49.
In contrast, the terminase C-terminal domains are more divergent and show little sequence homology. This may reflect functional differences between the various terminase enzymes –those that cleave concatemeric DNA at discrete sites and package a unit-length genome (e.g., lambda) vs. those that have a “non-specific” nuclease activity and package DNA via a head-full mechanism (e.g., T4). In addition, these domains must recognize specific sequences in viral DNA to initiate the packaging process. It is therefore not surprising that they diverge at the primary sequence level, commensurate with their role in maturation of specific viral DNA. That said, the one feature that is common to all of these proteins is nuclease activity and it is of interest that the terminase DNA maturation domains share sequence similarities to the Holliday junction resolvase RuvC and to a lesser extent the HIV integrase domains50.
The DNA Maturation ATPase Site (WA2)
Kinetic analysis of terminase holoenzyme identified a high-affinity ATPase activity in gpA (Km,app~ 5 μM)11; 36; 37. Further, kinetic analysis of the nuclease, strand-separation and packaging activities of the holoenzyme showed that each reaction is maximally stimulated by low ATP concentrations (1 – 50 μM)24; 26; 27; 31; 37. This has been interpreted to indicate that gpA possesses a single (high affinity) ATPase site that is involved in both DNA maturation and DNA packaging reactions. The alternate explanation, that gpA possesses multiple ATPase sites with similar nucleotide binding affinities, could not be ruled out, however.
The present work clearly demonstrates that the DNA maturation domain contains a discrete ATP binding and hydrolysis site, presumably identified by the Walker A sequence centered at Lys497 (WA2, Figure 2A)33. This residue represents the conserved lysine that interacts with the γ-phosphate of ATP (or GTP) and plays a major role in nucleotide binding and/or hydrolysis in all characterized P-loops34. While alteration of gpA-K497 only modestly affects ATP binding, mutation of this residue strongly attenuates ATP hydrolysis, consistent with a role as a conserved P-loop residue. Moreover, mutation of this residue abrogates the nuclease and helicase activities of the protein (data shown here; 17). In sum, the data clearly establish that the gpA DNA maturation domain contains a dedicated ATPase site and that ATP is required for DNA maturation. We note that primary sequence analysis similarly identified an ATPase motif within the DNA maturation domain of the large T4 terminase subunit33; extensive mutagenesis of this region failed to expose this putative catalytic site, however51. This again may reflect functional differences between viruses like T4 that need only cleave the duplex to mature the ends and those like lambda that must also physically separate the nicked DNA strands, a reaction that requires the hydrolysis of ATP.
Biological Implications of a “Single-Turnover” Maturation ATPase
Maturation of the genome end requires separation of the nicked strands created by the nuclease activity of gpA, a reaction that is fueled by ATP hydrolysis15; 24. We note that terminase melts the 12 base-pairs of DNA within the nicked cosN site but does not further engage the duplex24. For his reason, we have previously suggested that this reaction is more appropriately described as a strand separation activity that mechanistically represents a single helicase catalytic turnover (see Figure 1)24; 52. A direct consequence is that a single round of ATP hydrolysis (a single catalytic turnover) may be all that is required to power the reaction. This predicts that steady-state turnover at the DNA maturation ATPase site will be quite limited, which is consistent with both the single-turnover and steady-state kinetic data reported here. Moreover, the observation that ADP binds to the protein more tightly than does ATP suggests that ADP release may be the rate-limiting step in steady state ATP hydrolysis at the WA2 site. Whatever the case, the “single-turnover” catalysis observed here is consistent with the observation that the DNA maturation ATPase site is “catalytically silent” in context of the full-length protein, effectively masked by ATP hydrolysis at the high affinity DNA packaging ATPase site.
We note here that previous single-turnover ATPase analysis of the isolated full-length gpA subunit revealed a bi-phasic time course, with fast and slow rates of 0.706 ± 0.089 min−1 and 0.017 ± 0.025 min−1, respectively42. A mechanistic interpretation for the observed bi-phasic kinetics was not possible at that time, however, as it was not appreciated that the protein contained two functional ATPase catalytic sites. The demonstration here that gpA contains a second ATPase catalytic site now allows appropriate interpretation. Importantly, single-turnover ATP hydrolysis by gpA-ΔN179 exhibits a mono-phasic time course with an observed rate of 0.086 ± 0.008 min−1, which closely approximates the slow phase rate constant observed with the full-length protein. We interpret these data to indicate that fast turnover occurs at the DNA packaging site while the slow rate represents ATP hydrolysis at the DNA maturation ATPase site.
Biological Implications of a “Low Affinity” Maturation ATPase
A major role for ATP hydrolysis by gpA is to fuel DNA packaging by terminase holoenzyme. This function is provided by the high affinity ATPase site situated in the N-terminal DNA packaging domain of gpA (WA1, Figure 2A)19; 21; 36. The genome end must be matured prior to packaging, however, a process that requires the nuclease and strand separation activities of the enzyme (Figure 1)9. It has long been appreciated that the nuclease activity of gpA is regulated by ATP binding and/or hydrolysis. We show here that a discrete, C-terminal DNA maturation ATPase site in the protein provides this role. Within this context, the disparate affinities of the two sites for ATP is of interest. The packaging ATPase has a Km~ 5 μM. Given that the concentration of ATP in vivo is approximately 2 mM53, this site is continuously saturated with ATP. This ensures that the “gas can” is always full and that active DNA packaging is not affected by fluctuations in the energy status of the cell. In contrast, the DNA maturation ATPase has a KD,app~ 2 mM, which is similar to the ATP concentration within the cell. This observation is harmonious with the concept that an enzyme possesses a Km similar to the in vivo concentration of the relevant ligand, which allows regulation of enzyme activity by the metabolic status of the cell. Based on this concept, we propose that the energy status of the E. coli cell may regulate virus assembly, as follows. When ATP is plentiful, the ATP-dependent nuclease activity of gpA is efficient and maturation of the genome proceeds rapidly. As the concentration of ATP drops, packaging initiation is attenuated. We note that the gpNu1 subunit, whose presumed role is to assemble the packaging machinery specifically at cos, has a Km~ 0.5 – 1 mM for ATP. Thus, ATP may regulate packaging initiation at two levels – (i) the initial assembly of the packaging machinery at cos (regulated by the gpNu1 ATPase) and (ii) maturation of the genome end (regulated by a gpA ATPase). Whatever the case, processive DNA packaging proceeds efficiently once the process has been initiated due to the high affinity of the packaging ATPase for ATP. We note that Serwer has proposed a similar ATP-regulated packaging model for bacteriophage T3/T7, though the mechanistic details of the model differ from ours54.
Lys497 and the DNA Maturation Activities of gpA
Feiss and co-workers previously reported that mutation of gpA-K497 abrogated the nuclease and helicase activities of terminase holoenzyme17. The data presented here confirm this observation and further show that decreased activity is not a consequence of decreased DNA binding affinity. The simplest explanation is that Lys497 directly participates in the chemical step of the nuclease reaction. We disfavor this concept, however, because it is clear that this residue is directly involved in ATP hydrolysis and it is unlikely that the identical residue would also directly participate in the duplex nicking reaction. Rather, these studies suggest that ATP hydrolysis serves as a conformational switch, linking the ATP hydrolytic cycle to changes in protein conformation that modulate both the nuclease and strand-separation activities of the enzyme. This model finds mechanistic similarity to the nucleotide regulated activities of a diverse group of proteins, including the hsp60/hsp70 molecular chaperones55, ATPase p97 (an ATPase that plays a role in organelle membrane fusion reactions)56, ClpB (ATP-dependent molecular chaparone)57, DNA gyrase and the MutL DNA mismatch repair protein58; 59, among others60. A common mechanistic theme emerges where ATP binding and/or hydrolysis trigger conformational changes that can be quite dramatic and that perform distinct functions in the catalytic cycle of the enzyme complex.
Lambda terminase holoenzyme is a complex biological machine that contains a nuclease catalytic site, a strand-separation “helicase” catalytic site, a DNA translocation catalytic site and four ATPase sites (two in gpA and one in each of the gpNu1 dimer subunits). Each of these catalytic sites must be regulated both spatially and temporally to ensure site-specific assembly of the DNA maturation machinery, appropriate maturation of the genome end, binding to the procapsid and activation of DNA translocation motor. Complex allosteric interactions between all of the catalytic sites have been demonstrated9; 37; 61 which likely reflects the regulated activation and de-activation of the sites. Given the similarities among all of the viral packaging motors, lessons learned in the lambda system will likely have broad biological implications. Biochemical, biophysical and structural studies currently underway in our lab seek to dissect these intricate allosteric interactions and to define the mechanistic details of DNA maturation and DNA packaging by these fascinating molecular motor complexes.
Materials and Methods
Tryptone, yeast extract and agar were purchased from DIFCO. Restriction endonuclease enzymes, the Klenow fragment and mature λ DNA were purchased from Invitrogen. Chymotrypsin, trypsin, Endo-Glu-C (sequencing grade) and proteinase K were purchased from Sigma. Radionucleotides, chromatography resins (DEAE-sepharose FF, SP-sepharose FF and S-300 HR) and Superose 6 10/300 GL gel filtration columns were purchased from Amersham Bioscience. All of the synthetic oligonucleotides used in these studies were purchased from Invitrogen and were used without further purification. All other materials were of the highest quality commercially available.
Bacterial cultures were grown in shaker flasks utilizing a New Brunswick Scientific series 25 incubator/shaker. All protein purifications utilized a Pharmacia FPLC system, which consisted of two P500 pumps, a GP250-plus controller, a V7 injector and a Uvicord SII variable wavelength detector. UV-VIS absorbance spectra were recorded on a Hewlett-Packard HP8452A spectrophotometer.
Limited Proteolysis
Full-length gpA (20 μg) was subjected to limited proteolysis either with chymotrypsin, trypsin, Endo-Glu-C or proteinase K. The digestion buffer for chymotrypsin and trypsin was composed of 20 mM HEPES, pH 8.0, 50 mM NaCl, 2.5 mM MgCl2 and 2mM DTT and used a gpA:protease mass ratio of 200:1 and 400:1, respectively. The digestion buffer for Endo-Glu-C and proteinase K was composed of 10 mM Tris, pH 7.8 (room temperature), 5 mM EDTA and 0.5% SDS and used a gpA:protease mass ratio of 300:1 and 850:1, respectively. The reaction mixtures were maintained at room temperature and aliquots (10 μL) were removed at 5, 15, 30, 45, 60 and 90 minutes. The protease reaction was stopped with the addition of SDS-PAGE loading buffer and boiling for ten minutes, and the samples were analyzed by 15% SDS-PAGE to determine the appearance of protease resistant fragments.
N-Terminal Amino Acid Sequencing
GpA (20 μg) was digested with Endo-Glu-C and the reaction was stopped after 90 minutes as described above. The proteolytic fragments were fractionated by 15% SDS-PAGE and then electro-eluted from the gel onto a ProBlott® membrane (purchased from Applied Biosystems). Protease resistant fragments were selected and the appropriate bands were excised from the membrane and placed in the chamber of the sequenator for N-terminal sequence analysis. Sequence analysis was performed by the Macromolecular Resource Center at the University of Colorado Health Sciences Center.
Mass Spectrometry
GpA (20 μg) was digested with Endo-Glu-C and the reaction was stopped after 90 minutes as described above. The sample was directly submitted to MALDI-TOF mass spectrometry analysis, which was performed by the Macromolecular Resource Center at the University of Colorado Health Sciences Center.
Construction of gpA Deletion Mutants
Five Endo-Glu-C protease-resistant fragments were chosen for further analysis by cloning and over-expression in E. coli (see Figure 2). Truncated gpA genes were amplified by PCR using mature λ DNA as a template. The primers used were as follows:
| Forward Primers | EcoRI restriction sequence and the initiating methionine codon are indicated in underline and bold, respectively. |
| Met1 primer | 5′-GGGGGAATTCATGAATATATCGAACAGTC-3′ |
| Leu180 primer | 5′-GGGGGAATTCATGCTTGCTGCTTTTGATGAT-3 |
| Ala557 primer | 5′-GGGGGAATTCATGGCGCAGCAGCTGACT-3′ |
| Reverse Primers | HindIII restriction sequence and the stop codon are indicated in underline and bold, respectively |
| Glu641 Primer | 5′-GGGGGGATCCTCATTCATCCTCTCCGGATAA-3 |
| Glu556 primer | 5′-GGGGGGATCCTCATTCGGTCAGATCAAAAATA-3 |
| Glu179 primer | 5′-GGGGGGATCCTCAATGTTCATCATAACCCGCCA-3′ |
PCR conditions were as follows, (94°C for one minute, 50°C for one minute, and 72°C for two minutes) for 35 cycles. PCR products were purified and isolated using a Qiagen Gel Extraction Kit according to manufacturer protocol. The PCR fragments were digested with EcoRI and HindIII, cloned into similarly digested pKKT7, and used to transfect E. coli BL21(DE3) cells as described previously46.
Protein Expression
BL21(DE3) cells transformed with the relevant expression vector were grown to an O.D. of 0.6 (600 nm) at 37°C and protein expression was then induced with 0.4 mM IPTG. The temperature was reduced to 30°C and cell culture was continued for an additional three hours. The cells were harvested by centrifugation and the cell pellet was resuspended in 100 mL ice-cold Buffer A (20 mM Tris, pH 8.0 (4°C), 100 mM NaCl, 7 mM β-ME, 1 mM EDTA) containing 10% glycerol. Unless otherwise indicated, all subsequent steps were performed with ice-cold buffers at 0 to 4°C. The cells were lysed by sonication (fifteen 10 second bursts), insoluble material was removed by centrifugation (5000 x g x 10 minutes) and the pellet and supernatant fractions were examined for the presence of the desired construct by 15% SDS-PAGE.
Protein Purification
Proteins in the crude cell lysis supernatant were sequentially fractionated with 10% and 50% saturated ammonium sulfate. The 50% pellet was taken into 20 mL Buffer A and dialyzed against the one liter of buffer A overnight. The sample was then applied to a DEAE sepharose column (200 mL) and eluted with a salt gradient to 550 mM NaCl. The column fractions (15 mL) were examined for the presence of the desired protein by SDS-PAGE and the appropriate fractions were pooled and dialyzed against Buffer A.
The sample was next loaded onto a SP-sepharose column (20 mL) and the proteins were eluted with a salt gradient to 400 mM NaCl. The column fractions (12 mL) were analyzed for the presence of the desired protein. Appropriate fractions were pooled and dialyzed against Buffer A. The pooled SP-sepharose fractions were next loaded onto a pre-packed Sephacryl S-300 gel filtration column (125 mL), which was developed with Buffer A at a flow rate of 0.5 mL/min. Elution fractions (10 mL) were analyzed for the presence of protein and desired fractions were pooled. The purified protein preparations were concentrated using Amicon® concentration devices and stored in Buffer A at −80°C. All protein preparations were greater than 95% homogenous as determined by SDS-PAGE and densitometer analysis as previously described10. The concentration of each protein was determined spectrally by the method of Gill and von Hippel62.
Analytical Gel Filtration
The protein sample was applied to a Superose 6 10/300 GL gel filtration column equilibrated with Buffer A and the proteins were eluted with the same buffer at a flow-rate of 0.2 mL/minute. The column was calibrated with the following molecular weight standards: blue dextran (FW> 2,000 kDa), ferritin (440 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa) and ribonuclease A (13.7 kDa).
Fluorescence Spectroscopy
All fluorescence spectra were recorded on a Photon Technologies Quanta-Master spectrofluorometer equipped with a Masterline-Forma model 2095 circulating water bath and a thermostated cell holder maintained at 25°C. Spectra were recorded using a protein concentration of 1 μM in Buffer A with constant stirring. Fluorescence emission spectra were recorded from 320 nm to 380 nm using a λex= 295 nm, a slit width of 2 nm, a step size of 1 nm and an integration time of 5 seconds.
Fluorescence-Monitored Ligand Binding
Nucleotide binding to the constructs was monitored by changes in intrinsic tryptophan fluorescence of the protein as a function of increasing nucleotide concentration. Fluorescence emission spectra were corrected for background, dilution and inner filter effects using61:
| (1) |
where Fcorr is the corrected fluorescence intensity, Fo is the observed fluorescence of the sample, FB is the background fluorescence intensity of the buffer, vi and vf are the initial volume and final volumes, respectively, of the sample after addition of the titrant, b the path length of the cell, and Aem and Aex are the absorbance values of the ligand(s) at the emission and excitation wavelengths, respectively, at each point in the titration. The binding data were fit to a simple Langmuir binding model using:
| (2) |
where Qobs represents percent quenching of the intrinsic protein fluorescence intensity observed in the presence of ligand at concentration [N], Qmax represents quenching at infinite ligand concentration, KD,app is the apparent equilibrium association constant for the interaction and b is the baseline offset for the titration data.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra were recorded on an Aviv model 62DS circular dichroism spectrophotometer equipped with a Brinkman Lauda RM6 circulating water bath and a thermostated cell holder. Data were collected at 4°C in Buffer A. Far-UV CD spectra were recorded from 200 nm – 260 nm using a protein concentration of 0.1 mg/mL in a 0.1 cm strain-free quartz cuvette. Near-UV CD spectra were recorded from 250 nm – 350 nm using a protein concentration of 1.0 mg/mL in a 0.1 cm strain-free quartz cuvette. In both cases, data were collected at 0.5 nm intervals using a bandwidth of 1.5 nm and dwell times of 30 seconds. The raw spectra were converted to molar ellipticity using the following equation:
| (3) |
where θ is the molar ellipticity (degrees-cm2/dmol), θobs is the ellipticity recorded by the instrument (mdeg), MRW is the mean residue weight, b is the cell path length in cm and c is the protein concentration in mg/mL. Protein secondary structure content was determined by analysis of the far-UV CD data as previously described44. CD-monitored thermally-induced protein unfolding experiments were performed and analyzed as previously described46.
Electrophoretic Mobility Shift Experiments
DNA binding experiments were performed in buffer containing 20 mM Tris, pH 8.0 (4°C), 50 mM NaCl, 2 mM spermidine, 1 mM EDTA, 7 mM β-ME and 10% glycerol. The specific DNA binding substrate was cos-DNA, a 272 bp duplex that contains the entire λ cos sequence, while the non-specific DNA binding substrate was ns-DNA, a 272 base-pair duplex of random sequence63. Radiolabeled DNA was included at a concentration of 20 pM and herring sperm DNA (100 pM) was added to all binding mixtures. Protein was added as indicated in each individual experiment and the binding mixture was incubated at 25°C for 15 minutes. The mixture was then loaded onto a 5% polyacrylamide gel (acrylamide:bis-acrylamide ratio of 80:1) and run at 15 V/cm in 0.5X TBE at 4°C for 1.5 hours. The gel was then dried in vacuo on Whatmann 3MM filter paper and the radioactive bands were visualized and quantified using a Molecular Dynamics Storm system and the Molecular Dynamics ImageQuant® data analysis package as previously described25; 63.
ATPase Activity Assay
Steady-state ATPase assays (20 μL) used a buffer composed of 50 mM Tris, pH 9.0 (4°C), 10 mM MgCl2, 2 mM α32P-ATP, 1.5 nM lambda DNA, 2 mM spermidine and 7 mM β-ME. The reaction was initiated with the addition of protein (2 μM) and the mixture was incubated at 37°C for 30 minutes. The reaction was stopped with the addition of 20 °L quench buffer (5 mM ATP, 5 mM ADP and 100 mM EDTA), the nucleotides were fractionated by thin layer chromatography and the hydrolysis of ATP quantified by phosphor image analysis as previously described11.
Single-turnover ATPase assays were conducted similarly except that ATP and protein concentrations were 0.1 μM and 17.5 μM, respectively, and aliquots of the reaction mixture were removed and quenched at the indicated time points. The data were analyzed according to the equation:
where [ADP]τ refers to the ADP formed at time τ, A is the extent of the reaction at τ= ∞ and k represent the observed rate constant. The indicated constants were determined by non-linear regression analysis of the experimental data using the Igor® data analysis program (Wave Metrics, Lake Oswego, OR) as described previously 42.
Endonuclease Activity Assay
The cos-cleavage endonuclease assay was performed using pAFP1 as the DNA substrate as previously described16. Briefly, the reactions mixtures were composed of 20 mM Tris, pH 8.0 (4°C), 10 mM MgCl2, 2 mM ATP, 7 mM β-ME, 2 mM spermidine and 20 nM linearized pAFP1 DNA substrate. The reaction was initiated with the addition of protein (2 μM) and the reaction mixtures were incubated at 37°C for 20 minutes. The reaction was stopped with the addition of 2 μL quench buffer (200 mM EDTA, 20% glycerol, 0.16% bromphenol blue and 0.16% xylene cyanol) and the DNA products were fractionated on a 0.8% agarose gel. The reaction products were analyzed by video densitometry as previously described16.
Supplementary Material
Acknowledgments
The authors wish to express their gratitude to Drs. Michael Feiss and Nasib Karl Maluf for their helpful discussions and essential review of this manuscript. This work was supported by the National Science Foundation Grant MCB-0111066.
This work was supported by National Science Foundation Grant MCB-0111066.
Footnotes
Abbreviations used. cos, the junction between individual genomes in concatemeric λ DNA and the site where terminase assembles to initiate the packaging process; cosN, a subsite within the cos sequence where the terminase gpA subunit introduces symmetric nicks, twelve bases apart to mature concatemeric λ DNA; gpA-FL, the isolated full-length large terminase subunit; gpA-ΔN179, a gpA construct in which the N-terminal 179 residues have been deleted; gpA-ΔN179,K497D, the gpA-ΔN179 deletion construct into which the lysine at position 497 has mutated to aspartic acid; immature DNA, a DNA concatemer consisting of multiple λ genomes covalently linked in a head-to-tail fashion; mature DNA, an individual λ genome that is excised from the concatemer and that is found within the capsid of an infectious virus particle; terminase holoenzyme, the functional λ terminase enzyme composed of gpA and gpNu1 subunits in a gpA1●gpNu12 protomer complex; WA1, the N-terminal Walker A (P-loop) motif associated with the DNA packaging activity of λ terminase; WA2, the C-terminal Walker A (P-loop) motif associated with the DNA maturation (nuclease/helicase) activity of λ terminase.
For simplicity, we assume a minimal kinetic mechanism where nucleotide binding is directly followed by phosphodiester bond hydrolysis. We note, however, that conformational change steps preceding the chemical step will also contribute to the observed single-turnover rate.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Casjens SR. An Introduction to Virus Structure and Assembly. In: Casjens SR, editor. Virus Structure and Assembly. Jones and Bartlett Publishers, Inc; Boston, MA: 1985. pp. 1–28. [Google Scholar]
- 2.Black LW. DNA Packaging in dsDNA Bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 3.Catalano CE. Viral Genome Packaging Machines: An Overview. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Landes Bioscience/Eurekah.com; Georgetown, TX: 2005. pp. 1–4. [Google Scholar]
- 4.Fields BN, Knipe DM, Howley PM. Fields Virology. 3. Two vols, Lippincott-Raven Publishers; Philadelphia, PA: 1996. [Google Scholar]
- 5.Fujisawa H, Morita M. Phage DNA Packaging. Genes to Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 6.Murialdo H. Bacteriophage Lambda DNA Maturation and Packaging. Annu Rev Biochem. 1991;60:125–153. doi: 10.1146/annurev.bi.60.070191.001013. [DOI] [PubMed] [Google Scholar]
- 7.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of Bacteriophage Lambda Terminase into a Viral DNA Maturation and Packaging Machine. Biochemistry. 2006 doi: 10.1021/bi0615036. in press. [DOI] [PubMed] [Google Scholar]
- 8.Maluf NK, Yang Q, Catalano CE. Self-association Properties of the Bacteriophage [lambda] Terminase Holoenzyme: Implications for the DNA Packaging Motor. J Mol Biol. 2005;347:523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Feiss M, Catalano CE. Bacteriophage Lambda Terminase and the Mechanism of Viral DNA Packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Landes Bioscience/Eurekah.com; Georgetown, TX: 2005. pp. 5–39. [Google Scholar]
- 10.Tomka MA, Catalano CE. Physical and Kinetic Characterization of the DNA Packaging Enzyme from Bacteriophage Lambda. J Biol Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 11.Tomka MA, Catalano CE. Kinetic Characterization of the ATPase Activity of the DNA Packaging Enzyme from Bacteriophage Lambda. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 12.Rubinchik S, Parris W, Gold M. The in Vitro ATPases of Bacteriophage Lambda Terminase and Its Large Subunit, Gene Product A. J Biol Chem. 1994;269:13586–13593. [PubMed] [Google Scholar]
- 13.Rubinchik S, Parris W, Gold M. The in Vitro Endonuclease Activity of Gene Product A, the Large Subunit of Bacteriophage Lambda Terminase, and Its Relationship to the Endonuclease Activity of the Holoenzyme. J Biol Chem. 1994;269:13575–13585. [PubMed] [Google Scholar]
- 14.Rubinchik S, Parris W, Gold M. The in Vitro Translocase Activity of Lambda Terminase and Its Subunits. J Biol Chem. 1995;270:20059–20066. doi: 10.1074/jbc.270.34.20059. [DOI] [PubMed] [Google Scholar]
- 15.Parris W, Rubinchik S, Yang YC, Gold M. A New Procedure for the Purification of the Bacteriophage Lambda Terminase Enzyme and Its Subunits. J Biol Chem. 1994;269:13564–13574. [PubMed] [Google Scholar]
- 16.Woods L, Terpening C, Catalano CE. Kinetic Analysis of the Endonuclease Activity of Phage Lambda Terminase: Assembly of a Catalytically-Competent Nicking Complex is Rate-Limiting. Biochemistry. 1997;36:5777–5785. doi: 10.1021/bi963044m. [DOI] [PubMed] [Google Scholar]
- 17.Hwang Y, Feiss M. Mutations Affecting the High Affinity ATPase Center of gpA, the Large Subunit of Bacteriophage Lambda Terminase, Inactivate the Endonuclease Activity of Terminase. J Mol Biol. 1996;261:524–535. doi: 10.1006/jmbi.1996.0480. [DOI] [PubMed] [Google Scholar]
- 18.Hang JQ, Tack BF, Feiss M. ATPase Center of Bacteriophage l Terminase Involved in Post-Cleavage Stages of DNA Packaging: Indentification of ATP-Interactive Amino Acids. J Mol Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- 19.Duffy C, Feiss M. The Large Subunit of Bacteriophage Lambda’s Terminase Plays a Role in DNA Translocation and Packaging Termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 20.Davidson AR, Gold M. Mutations Abolishing the Endonclease Activity of Bacteriophage Lambda Terminase Lie in Two Distinct Regions of the A Gene, One of which May Encode a “Leucine Zipper” DNA-Binding Domain. Virol. 1992;189:21–30. doi: 10.1016/0042-6822(92)90677-h. [DOI] [PubMed] [Google Scholar]
- 21.Dhar A, Feiss M. Bacteriophage Lambda Terminase: Alterations of the High-affinity ATPase Affect Viral DNA Packaging. J Mol Biol. 2005;347:71–80. doi: 10.1016/j.jmb.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Bear S, Court D, Friedman D. An Accessory Role for Escherichia coli Integration Host Factor: Characterization of a Lambda Mutant Dependent Upon Integration Host Factor for DNA Packaging. J Virol. 1984;52:966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinder G, Gold M. The Nu1 Subunit of Bacteriophage Lambda Terminase Binds to Specific Sites in cos DNA. J Virol. 1988;62:387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q, Catalano CE. Kinetic Characterization of the Helicase Activity of the DNA Packaging Enzyme from Bacteriophage Lambda. Biochemistry. 1997;36:10638–10645. doi: 10.1021/bi970689t. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Hanagan A, Catalano CE. Assembly of a Nucleoprotein Complex Required for DNA Packaging by Bacteriophage Lambda. Biochemistry. 1997;36:2744–2752. doi: 10.1021/bi9622682. [DOI] [PubMed] [Google Scholar]
- 26.Yang Q, Catalano CE. Biochemical Characterization of Bacteriophage Lambda Genome Packaging in Vitro. Virology. 2003;305:276–287. doi: 10.1006/viro.2002.1602. [DOI] [PubMed] [Google Scholar]
- 27.Hwang Y, Feiss M. A Defined System for in Vitro Lambda DNA Packaging. Virology. 1995;211:367–376. doi: 10.1006/viro.1995.1419. [DOI] [PubMed] [Google Scholar]
- 28.Goodrich JA, Tjian R. Transcription Factors IIE and IIH and ATP Hydrolysis Direct Promoter Clearance by RNA Polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 29.Catalano CE. The Terminase Enzyme from Bacteriophage Lambda: A DNA-Packaging Machine. Cellular and Molecular Life Sciences. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins RR, Lucko HJ, Becker A. Mechanism of cos DNA Cleavage by Bacteriophage Lambda Terminase: Multiple Roles of ATP. Cell. 1988;54:765–775. doi: 10.1016/s0092-8674(88)91021-5. [DOI] [PubMed] [Google Scholar]
- 31.Gaussier H, Yang Q, Catalano CE. Building a Virus from Scratch: Assembly of an Infectious Virus Using Purified Protein Components in a Rigorously Defined Biochemical Assay System. J Mol Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Becker A, Gold M. Prediction of an ATP Reactive Center in the Small Subunit, gpNu1, of the Phage Lambda Terminase Enzyme. J Mol Biol. 1988;199:219–222. doi: 10.1016/0022-2836(88)90391-9. [DOI] [PubMed] [Google Scholar]
- 33.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-Dependent ATPase Activity of the DNA Packaging Protein gp16 of Bacteriophage ø29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 34.Saraste M, Sibbald PR, Wittinghofer A. The P-Loop, A Common Motif in ATP- and GTP-Binding Proteins. TIBS. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 35.Vetter I, Wittinghofer A. Nucleoside Triphosphate-Binding Proteins: Different Scaffolds to Achieve Phosphoryl Transfer. Quart Rev Biophys. 1999;32:1–56. doi: 10.1017/s0033583599003480. [DOI] [PubMed] [Google Scholar]
- 36.Hwang Y, Catalano CE, Feiss M. Kinetic and Mutational Dissection of the Two ATPase Activities of Terminase, the DNA Packaging Enzyme of Bacteriophage Lambda. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, Catalano CE. A Minimal Kinetic Mechanism for a Viral DNA Packaging Machine. Biochemistry. 2004;43:289–299. doi: 10.1021/bi035532h. [DOI] [PubMed] [Google Scholar]
- 38.Babbar BK, Gold M. ATP-Reactive Sites in the Bacteriophage Lambda Packaging Protein Terminase Lie in the N-Termini of its Subunits, gpA and gpNu1. Virology. 1998;247:251–264. doi: 10.1006/viro.1998.9221. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence Analysis of Bacteriophage T4 DNA Packaging/Terminase Genes 16 and 17 Reveals a Common ATPase Center in the Large Subunit of Viral Terminase. Nuc Acid Res. 2002;30:4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, de Beer T, Woods L, Meyer J, Manning M, Overduin M, Catalano CE. Cloning, Expression, and Characterization of a DNA Binding Domain of gpNu1, a Phage Lambda DNA Packaging Protein. Biochemistry. 1999;38:465–477. doi: 10.1021/bi981271d. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q, Berton N, Manning MC, Catalano CE. Domain Structure of gpNu1, a Phage Lambda DNA Packaging Protein: Identification of Self-Association and gpA-Interactive Domains. Biochemistry. 1999;38:14238–14247. doi: 10.1021/bi991408f. [DOI] [PubMed] [Google Scholar]
- 42.Hang JQ, Woods L, Feiss M, Catalano CE. Cloning, Expression, and Biochemical Characterization of HexaHistidine-Tagged Terminase Proteins. J Biol Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
- 43.de Beer T, Meyer J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into Specific DNA Recognition During the Assembly of a Viral Genome Packaging Machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 44.Bain DL, Berton N, Ortega M, Baran J, Yang Q, Catalano CE. Biophysical Characterization of the DNA Binding Domain of gpNu1, a Viral DNA Packaging Protein. J Biol Chem. 2001;276:20175–20181. doi: 10.1074/jbc.M100517200. [DOI] [PubMed] [Google Scholar]
- 45.Wu WF, Christiansen S, Feiss M. Domains for Protein-Protein Interactions at the N and C Termini of the Large Subunit of Bacteriophage Lambda Terminase. Genetics. 1988;119:477–484. doi: 10.1093/genetics/119.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer JD, Hanagan A, Manning MC, Catalano CE. The Phage Lambda Terminase Enzyme: 1. Reconstitution of the Holoenzyme from the Individual Subunits Enhances the Thermal Stability of the Small Subunit. Int J Biol Macromol. 1998;23:27–36. doi: 10.1016/s0141-8130(98)00009-9. [DOI] [PubMed] [Google Scholar]
- 47.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The Functional Domains of Bacteriophage T4 Terminase. J Biol Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 48.Story RM, Li H, Abelson JN. Crystal Structure of a DEAD Box Protein from the Hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci, USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun S, Kondabagil K, Gentz PM, Fossmann MG, Rao VB. The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Capsids. Mol Cell. 2007;23:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Ponchon L, Boulanger P, Labesse G, Letellier L. The endonuclease domain of bacteriophage terminases belongs to the resolvase/integrase/ribonuclease H superfamily: a bioinformatics analysis validated by a functional study on bacteriophage T5. J Biol Chem. 2006;281:5829–5836. doi: 10.1074/jbc.M511817200. [DOI] [PubMed] [Google Scholar]
- 51.Rentas FJ, Rao VB. Defining the Bacteriophage T4 DNA Packaging Machine: Evidence for a C-terminal DNA Cleavage Domain in the Large Terminase/Packaging Protein gp17. J Mol Biol. 2003;334:37–52. doi: 10.1016/j.jmb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 52.Lohman TM. Helicase-Catalyzed DNA Unwinding. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 53.Neidhardt FC. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Vol. 1. ASM Press; Washington, D.C: 1987. [Google Scholar]
- 54.Sun M, Louie D, Serwer P. Single-Event Analysis of the Packaging of Bacteriophage T7 DNA Concatemers in Vitro. Biophys J. 1999;77:1627–1637. doi: 10.1016/S0006-3495(99)77011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bukau B, Weissman J, Horwich A. Molecular Chaperones and Protein Quality Control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 56.Rouiller I, Butel VM, Latterich M, Milligan RA, Wilson-Kubalek EM. A Major Conformational Change in p97 AAA ATPase upon ATP Binding. Mol Cell. 2000;6:1485–1490. doi: 10.1016/s1097-2765(00)00144-1. [DOI] [PubMed] [Google Scholar]
- 57.Lee S, Choi JM, Tsai FTF. Visualizing the ATPase Cycle in a Protein Disaggregating Machine: Structural Basis for Substrate Binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends in Biochemical Sciences. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 60.Ogura T, Wilkinson AJ. AAA+ Superfamily ATPases: Common Structure -Diverse Function. Genes to Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 61.Gaussier H, Ortega ME, Maluf NK, Catalano CE. Nucleotides Regulate the Conformational State of the Small Terminase Subunit from Bacteriophage Lambda: Implications for the Assembly of a Viral Genome-Packaging Motor. Biochemistry. 2005;44:9645–9656. doi: 10.1021/bi050333e. [DOI] [PubMed] [Google Scholar]
- 62.Gill SC, von Hippel PH. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 63.Ortega ME, Catalano CE. Bacteriophage Lambda gpNu1 and Escherichia coli IHF Proteins Cooperatively Bind and Bend Viral DNA: Implications for the Assembly of a Genome-Packaging Motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.