Abstract
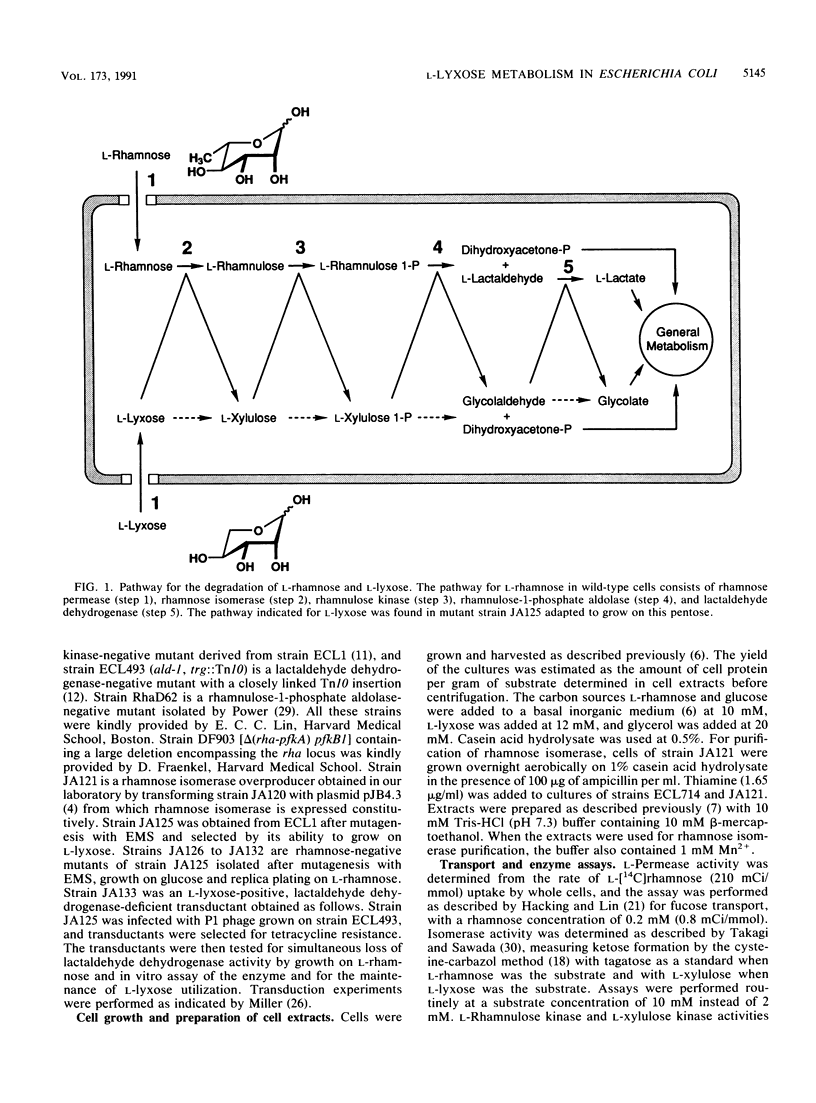
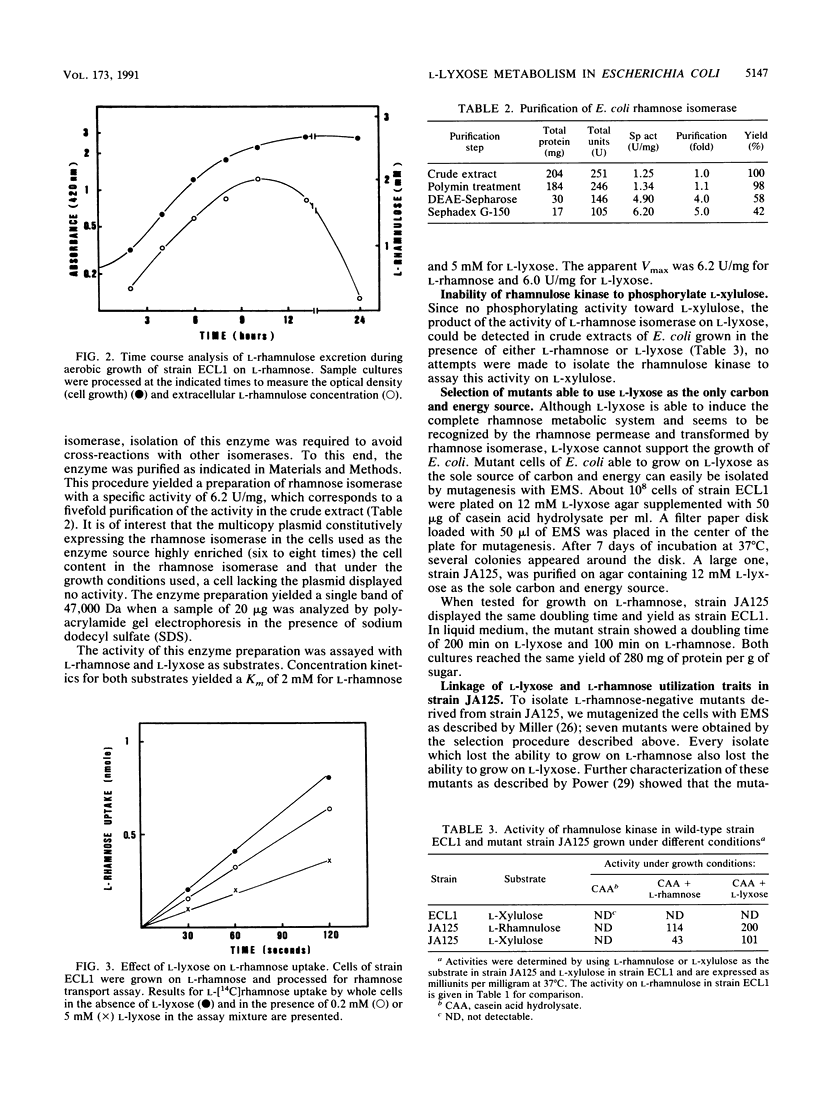
Escherichia coli cannot grow on L-lyxose, a pentose analog of the 6-deoxyhexose L-rhamnose, which supports the growth of this and other enteric bacteria. L-Rhamnose is metabolized in E. coli by a system that consists of a rhamnose permease, rhamnose isomerase, rhamnulose kinase, and rhamnulose-1-phosphate aldolase, which yields the degradation products dihydroxyacetone phosphate and L-lactaldehyde. This aldehyde is oxidized to L-lactate by lactaldehyde dehydrogenase. All enzymes of the rhamnose system were found to be inducible not only by L-rhamnose but also by L-lyxose. L-Lyxose competed with L-rhamnose for the rhamnose transport system, and purified rhamnose isomerase catalyzed the conversion of L-lyxose into L-xylulose. However, rhamnulose kinase did not phosphorylate L-xylulose sufficiently to support the growth of wild-type E. coli on L-lyxose. Mutants able to grow on L-lyxose were analyzed and found to have a mutated rhamnulose kinase which phosphorylated L-xylulose as efficiently as the wild-type enzyme phosphorylated L-rhamnulose. Thus, the mutated kinase, mapped in the rha locus, enabled the growth of the mutant cells on L-lyxose. The glycolaldehyde generated in the cleavage of L-xylulose 1-phosphate by the rhamnulose-1-phosphate aldolase was oxidized by lactaldehyde dehydrogenase to glycolate, a compound normally utilized by E. coli.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANDERSON R. L., WOOD W. A. Pathway of L-xylose and L-lyxose degradation in Aerobacter aerogenes. J Biol Chem. 1962 Feb;237:296–303. [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badía J., Baldomà L., Aguilar J., Boronat A. Identification of the rhaA, rhaB and rhaD gene products from Escherichia coli K-12. FEMS Microbiol Lett. 1989 Dec;53(3):253–257. doi: 10.1016/0378-1097(89)90226-7. [DOI] [PubMed] [Google Scholar]
- Baldomà L., Aguilar J. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J Biol Chem. 1987 Oct 15;262(29):13991–13996. [PubMed] [Google Scholar]
- Boronat A., Aguilar J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J Bacteriol. 1981 Jul;147(1):181–185. doi: 10.1128/jb.147.1.181-185.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat A., Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol. 1979 Nov;140(2):320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat A., Caballero E., Aguilar J. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J Bacteriol. 1983 Jan;153(1):134–139. doi: 10.1128/jb.153.1.134-139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter J. R., Gielow W. O. Properties of D-arabinose isomerase purified from two strains of Escherichia coli. J Bacteriol. 1973 Feb;113(2):687–696. doi: 10.1128/jb.113.2.687-696.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIU T. H., FEINGOLD D. S. THE PURIFICATION AND PROPERTIES OF L-RHAMNULOKINASE. Biochim Biophys Acta. 1964 Dec 23;92:489–497. doi: 10.1016/0926-6569(64)90009-4. [DOI] [PubMed] [Google Scholar]
- Caballero E., Baldomá L., Ros J., Boronat A., Aguilar J. Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J Biol Chem. 1983 Jun 25;258(12):7788–7792. [PubMed] [Google Scholar]
- Chen Y. M., Tobin J. F., Zhu Y., Schleif R. F., Lin E. C. Cross-induction of the L-fucose system by L-rhamnose in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3712–3719. doi: 10.1128/jb.169.8.3712-3719.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Zhu Y., Lin E. C. NAD-linked aldehyde dehydrogenase for aerobic utilization of L-fucose and L-rhamnose by Escherichia coli. J Bacteriol. 1987 Jul;169(7):3289–3294. doi: 10.1128/jb.169.7.3289-3294.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T. H., Feingold D. S. L-rhamnulose 1-phosphate aldolase from Escherichia coli. Crystallization and properties. Biochemistry. 1969 Jan;8(1):98–108. doi: 10.1021/bi00829a015. [DOI] [PubMed] [Google Scholar]
- Cocks G. T., Aguilar T., Lin E. C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J Bacteriol. 1974 Apr;118(1):83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DISCHE Z., BORENFREUND E. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J Biol Chem. 1951 Oct;192(2):583–587. [PubMed] [Google Scholar]
- Elsinghorst E. A., Mortlock R. P. D-arabinose metabolism in Escherichia coli B: induction and cotransductional mapping of the L-fucose-D-arabinose pathway enzymes. J Bacteriol. 1988 Dec;170(12):5423–5432. doi: 10.1128/jb.170.12.5423-5432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D. F., Sartoris M., Chiu T. H., Feingold D. S. L-rhamnulose 1-phosphate-1. Methods Enzymol. 1975;41:91–94. doi: 10.1016/s0076-6879(75)41021-7. [DOI] [PubMed] [Google Scholar]
- Hacking A. J., Lin E. C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J Bacteriol. 1976 Jun;126(3):1166–1172. doi: 10.1128/jb.126.3.1166-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeBlanc D. J., Mortlock R. P. Metabolism of D-arabinose: a new pathway in Escherichia coli. J Bacteriol. 1971 Apr;106(1):90–96. doi: 10.1128/jb.106.1.90-96.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Power J. The L-rhamnose genetic system in Escherichia coli K-12. Genetics. 1967 Mar;55(3):557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAGI Y., SAWADA H. THE METABOLISM OF L-RHAMNOSE IN ESCHERICHIA COLI. I. L-RHAMNOSE ISOMERASE. Biochim Biophys Acta. 1964 Oct 23;92:10–17. doi: 10.1016/0926-6569(64)90263-9. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Lin E. C. An evolvant of Escherichia coli that employs the L-fucose pathway also for growth on L-galactose and D-arabinose. J Mol Evol. 1986;23(3):259–266. doi: 10.1007/BF02115582. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Lin E. C. Loss of aldehyde dehydrogenase in an Escherichia coli mutant selected for growth on the rare sugar L-galactose. J Bacteriol. 1987 Feb;169(2):785–789. doi: 10.1128/jb.169.2.785-789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



