Summary
Anaphase-promoting complex or cyclosome (APC/C) controls the metaphase-to-anaphase transition and mitosis exit by triggering the degradation of key cell cycle regulators such as securin and B-type cyclins. However, little is known about the functions of individual APC/C subunits and how they might regulate APC/C activity in space and time. Here, we report that two potential Cdk1 kinase phosphorylation sites are required for the chromosomal localisation of GFP::Cdc27 during mitosis. Either or both of the highly conserved proline residues in the Cdk1 phosphorylation consensus sequence motifs were mutated to alanine (Cdc27 P304A or P456A). The singly mutated fusion proteins, GFP::Cdc27P304A and GFP::Cdc27P456A, can still localise to mitotic chromosomes in a manner identical to wild-type GFP::Cdc27 and are functional in that they can rescue the phenotype of the cdc27L7123 mutant in vivo. However, when both of the Cdk1 phosphorylation sequence motifs were mutated, the resulting GFP::Cdc27P304A,P456A construct was not localised to the chromosomes during mitosis and was no longer functional, as it failed to rescue mutant phenotypes of the cdc27L7123 gene. High levels of cyclin B and cyclin A were detected in mutant third instar larvae brain samples compared with its wild-type control. These results show for the first time that the two potential Cdk1 phosphorylation sites on Drosophila Cdc27 are required for its chromosomal localisation during mitosis and imply that these localisations specific to Cdc27 are crucial for APC/C functions.
Keywords: Metaphase, Mitosis, APC/C, Cdc27, Cdk1, Phosphorylation, Drosophila
Introduction
The anaphase-promoting complex or cyclosome (APC/C) is a large ubiquitin-protein ligase that controls the metaphase-to-anaphase transition and mitosis exit by triggering the degradation of a number of important cell cycle regulators such as securin and B-type cyclins (Irniger et al., 1995; King et al., 1995; Michaelis et al., 1997; Sudakin et al., 1995). The APC/C contains at least 10 different, evolutionarily conserved components and Cdc27 is one of its core components (King et al., 1995; Lamb et al., 1994; Sikorski et al., 1990; Tugendreich et al., 1995). APC/C activities are regulated in space and time during mitosis (Baumer et al., 2000; Clute and Pines, 1999; Huang and Raff, 1999; Lim et al., 1998; Yeong et al., 2000) through interactions with the APC/C activators Fzy/Cdc20 and Fzr/Cdh1 (Baumer et al., 2000; Kramer et al., 1998; Raff et al., 2002; Shirayama et al., 1999; Visintin et al., 1997; Yeong et al., 2000) and the APC/C inhibitors Mad2 and BubR1 (Sudakin et al., 2001). However, evidence has accumulated to suggest that differential localisation of APC/C components might also play roles in this regulation. This has been reported in other systems; for instance, Apc1/Tsg24 in CHO cells (Jorgensen et al., 1998) and Cdc27 (Apc3) in HeLa cells (Acquaviva et al., 2004) are centromere localised. Both Apc1/Tsg24 and Apc3 can be detected bound to isolated mitotic chromosomes, but Cdc16 has not been detected in these assays (Jorgensen et al., 1998). Similarly, Drosophila Cdc27 is associated with mitotic chromosomes, but Cdc16 is not (Huang and Raff, 2002). In A. nidulans, the Cdc27 homologue BimA is concentrated on spindle pole bodies (Mirabito and Morris, 1993). More recently, in PtK and HeLa cells, Cdc27 has been detected at kinetochores during prophase where it persists until metaphase, and also on chromosome arms where it persists from the onset of mitosis until anaphase (Topper et al., 2002). However, little is known about the functions of individual subunits of APC/C and the regulatory mechanisms and functional significance of the association of cdc27 with chromosomes.
It has been reported previously that many components of APC/C are hyper-phosphorylated during mitosis and Cdk1 kinase or polo-like protein kinase (Plk) activities play important roles in these phosphorylations in Xenopus, clam and human and in both fission and budding yeasts (Golan et al., 2002; May et al., 2002; Patra and Dunphy, 1998; Rudner and Murray, 2000; Shteinberg et al., 1999; Topper et al., 2002; Yamada et al., 1997). It is generally believed that these modifications are required for APC/C activity and possibly modulate APC/C sensitivity to inhibition by the mitotic checkpoint (Peters et al., 1996; Sudakin et al., 2001). These kinases can be co-fractionated with isolated chromosomes in human cells as well as with Cdc27 (Topper et al., 2002). These data suggest that Cdk1 or Polo kinase activities might directly or indirectly contribute to regulate Cdc27 chromosomal localisation. It is important to determine the potential roles of these kinase activities in the regulation of the APC/C in space and time.
We report here that two potential Cdk1 kinase phosphorylation sites are required for chromosomal localisation of GFP::Cdc27 during mitosis. It appears that these phosphorylations are crucial for APC/C function.
Results
Cdc27 phosphorylation is required for its chromosomal localisation
We have previously shown that Drosophila Cdc27 is associated with mitotic chromosomes (Fig. 1A1 and 7, 2 and 8, white arrows), but Cdc16 is not (Huang and Raff, 2002) (Fig. 1B1 and 7, 2 and 8, white arrows). To test whether the phosphorylation status of Cdc27 during mitosis contributes to its chromosomal localisation, GFP::Cdc27 or GFP::Cdc16 transgenic embryos at nuclear division cycles 8-9 were arrested at metaphase by microinjection with colchicine, a well-used spindle checkpoint activator. Activation of the spindle checkpoint increases and sustains Cdk1 and Plk kinase activities (Campbell et al., 1995; van Vugt et al., 2001; Weinert, 1997). After colchicine treatment, condensed chromosomes were arrested in mitosis (Fig. 1A6,B6): a clear indication that the spindle checkpoint was activated. Arrested chromosomes were highly enriched with GFP::Cdc27 that was distributed throughout the entire length of the chromosome arms (Fig. 1A3,9, white arrows), in comparison to non-treated controls that show a more diffuse association of GFP::Cdc27 with chromosomes at metaphase (Fig. 1A7, white arrow), although a clear chromosome association is seen at anaphase (Fig. 1A2,8, white arrow). In addition, GFP::Cdc27 also strongly associates with nuclear envelope membrane (Fig. 1A1,7, bright open ring structure, open arrowhead). Identical colchicine treatment of transgenic GFP::Cdc16-expressing syncytial embryos did not result in localisation of GFP::Cdc16 to arrested chromosomes (Fig. 1B3,9, arrows indicated shadow regions). As mentioned above, Cdc16 is one of the APC/C components that shows no chromosomal association during mitosis (Huang and Raff, 2002) (Fig. 1B7,2,8). These results further support the notion that Cdc27 phosphorylation status and by inference, Cdk1 or Plk kinase activity might have an important role in Cdc27 chromosomal localisation.
Fig. 1.
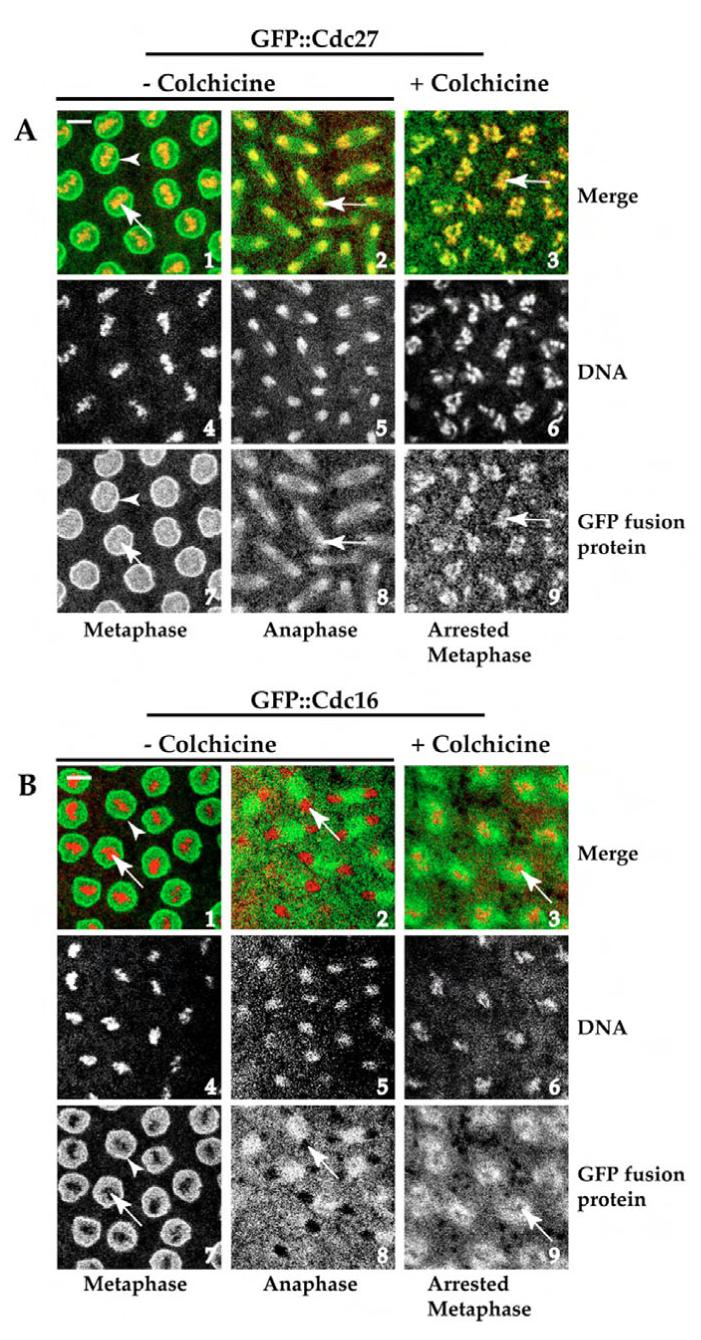
Confocal images showing small areas of Drosophila syncytial embryos that were taken from GFP::Cdc27 (A) or GFP::Cdc16 (B) transgenic flies with or without treatment with a 2.5 mM final intracellular concentration of colchicine. Drosophila GFP::Cdc27 associates with mitotic chromosomes (A, panels 1 and 7, 2 and 8, arrows), but GFP::Cdc16 does not (B, panels 1 and 7, 2 and 8, arrows indicate shadow regions), both GFP::Cdc27 and GFP::Cdc16 associate with nuclear envelope membrane (open arrowheads in A,B panels 1 and 7). Colchicine treatment clearly increases the association of GFP::Cdc27 with mitotic chromosomes (A, panels 3 and 9 arrows) compared with the metaphase control (A, panels 1 and 7, arrows). The localisation of the Drosophila GFP::Cdc16 is not affected by the same treatment (compare B, panels 3 and 9 with 1 and 7). A,B panels1-3: merge images (GFP::Cdc27 or GFP::Cdc16 in green, chromosomes in red), A,B panels 4-6: Rhodamine-H1-labelled chromosomes in white; A,B panels 7-9: GFP::Cdc27 or GFP::Cdc16 in white; A,B, panels 7 and 8: non-colchicine treated embryos; A,B, panel 9: colchicine-treated embryos. The developmental stage of syncytial embryos for each experiment was about nuclear division cycle 8 or 9. Bars, 10 μm.
There are two potential phosphorylation sites for Cdk1 protein kinase in the Drosophila Cdc27 amino acid sequence but there are none in Cdc16
Our findings suggest that phosphorylation is required for localisation of Cdc27 to chromosomes and that Cdk1 or Plk kinase activity play important roles in regulating this event. Cdk1, the cyclin-dependent kinase, is a highly Pro-directed kinase and readily phosphorylates S/TP sites in a number of mitotic substrates (Lew et al., 1992; Nigg, 1991; Shah et al., 2003; Shetty et al., 1993; Songyang et al., 1994). By contrast, Plk phosphorylation sites do not have a well-defined consensus sequence. We thus analysed the sequences of Cdc27 and Cdc16 using Scansite: (http://scansite.mit.edu/) Motif Scan search engine to identify potential consensus Cdk1 phosphorylation motifs. Two potential Cdk1 phosphorylation motifs were found in the Cdc27 sequence: T-303 (SSGTPFR) and S-456 (QPRSPPR) (Fig. 2A). For comparison, none were found in the Cdc16 sequence. To test whether Cdk1 phosphorylation is directly involved in regulation of Cdc27 chromosomal localisation, the two Cdk1 consensus phosphorylation motifs in Cdc27 were mutated either singly or together, replacing the essential Pro in the consensus sequence with Ala (Fig. 2A). This replacement completely abolishes phosphorylation by Cdk1 of the adjacent Ser or Thr residue (Lew et al., 1992). The corresponding DNA constructs (which have been fused with GFP at their 5′ ends) were used to generate transgenic flies.
Fig. 2.
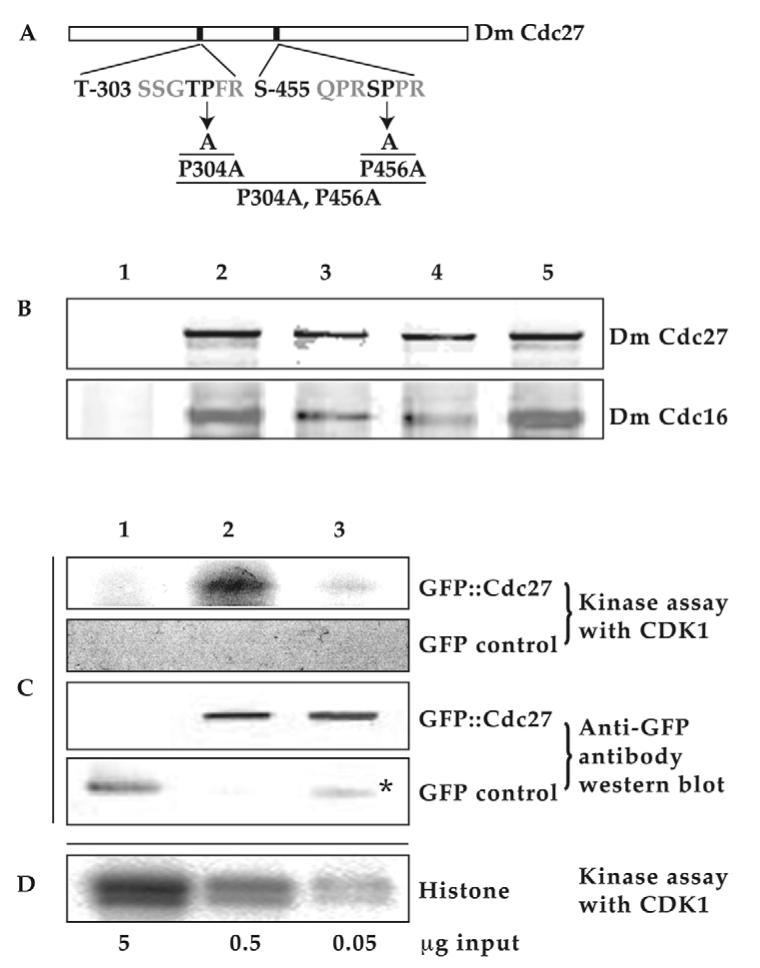
There are two potential phosphorylation sites for Cdk1 protein kinase in the Drosophila Cdc27 protein amino acid sequence. (A) Two consensus phosphorylation motifs for Cdk1 kinase (T-303 SSGTPFR and S-455 QPRSPPR) in the Drosophila Cdc27 amino acid sequence were identified using web site: http://scansite.mit.edu/. Highly conserved prolines (bold) from each motif were mutated to Alanine individually or together to create three mutated forms: P304A, P456A and double point mutations (P304A, P456A). All the constructs were fused with GFP at their N-terminus and used to generate transgenic Drosophila lines by the standard P-element-mediated transformation methods (see Materials and Methods). (B) Mutant Cdc27 proteins are still capable of incorporation into an APC/C complex. Embryo extracts made from control W67 or various GFP::Cdc27 fusion proteins as listed below, were immunoblotted with anti-Drosophila Cdc27 antibody (Top) or anti-Drosophila Cdc16 antibody (bottom) following co-immunoprecipitation (co-IP) of GFP fusion proteins with GFP antibody conjugated Dynabeads. Embryo extracts were made from an overnight collection of the flowing fly lines: lane 1, W67 (control); lane 2, gfp::cdc27/gfp::cdc27; lane 3, gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A; lane 4, gfp::cdc27P456A/gfp::cdc27P456A; lane 5, gfp::cdc27P304A/gfp::cdc27P304A. The amounts of immunoprecipitated mutant proteins were between 91-97% of those immunoprecipitated in wild-type controls. (C) Two potential Cdk1 phosphorylation sites contribute to the phosphorylation of Cdc27 in vitro by Cdk1. Samples were made from transgenic syncytial embryos. Lane 1, GFP; lane 2, GFP::Cdc27; lane 3, GFP::Cdc27P304A,P456A. Asterisk indicates GFP breakdown products from GFP::Cdc27P304A,P456A fusions. (D) Histone 1 kinase assay with Cdk1/cyclin B kinase in vitro as positive control.
Cdk1 kinase assay and western blot results confirm the loss of the phosphorylation potential in the doubly mutated GFP::Cdc27 fusion protein extracted from relevant transgenic embryos (Fig. 2C). The activity of a commercially available CDK1/cyclin B kinase (Millipore) was tested in vitro with histone 1 substrate (Fig. 2D) before it was used to carry out the kinase assay with the samples that carried the doubly mutated or non-mutated GFP::Cdc27 fusion proteins. After standard CDK1/cyclin B treatment, phosphorylation signals can only be detected with wild-type GFP::Cdc27 as substrate (Fig. 2C lane 2) but not with the doubly mutated GFP::Cdc27 or GFP alone as substrates (Fig. 2C lane 1 or lane 3), which show no detectable signal above background. GFP antibody western blot data show the protein loadings (Fig. 2C). These results strongly suggest that the doubly mutated Cdc27 fusion protein can no longer be specifically phosphorylated by CDK1/cyclin B kinase in vitro.
Immunoprecipitation results also show that all these three mutated GFP fusion proteins can still co-immunoprecipitated from supernatants with Cdc16, another core component of the APC/C (Fig. 2B, bottom panel). It was found that these singly or doubly mutated GFP::Cdc27 fusion proteins can be immunoprecipitated together with endogenous Cdc16 at comparable levels to wild-type protein (after comparing and quantifying the western blot band densities between Cdc16 and Cdc27 in wild-type or mutation samples, Fig. 2B and data not shown). Thus the mutations of the fusion proteins at P304A, P456A either singly or together (P304A, P456A) have not caused any conformational changes that would prevent the incorporation of the mutant fusion proteins into the APC/C complex.
Both Cdk1 phosphorylation sites are required for localisation of Cdc27 to the chromosomes
Results from confocal time-lapse images taken from either of the transgenic lines carrying only one Cdk1 consensus site mutation (GFP::Cdc27P304A or GFP::Cdc27P456A) showed no significant dynamic localisation differences to the wild-type GFP::Cdc27 transgenic line. In all three lines, GFP::Cdc27 localised to chromosomes during mitosis (Fig. 3A,B). All three transgenic lines were also capable of rescuing the cdc27L7123 mutant phenotype (see Materials and Methods), as evidenced by western blot results demonstrating that our stable rescued transgenic lines can develop healthily supported by these three different GFP::Cdc27 fusion proteins and showed no subcellular localisation differences during cell cycle progression (Fig. 3A,B, white open arrows, arrowheads and double arrowheads and 4A lane 2 and 3) (Huang and Raff, 2002). However when transgenic embryos containing the gfp::cdc27 transgene mutated at both potential Cdk1 phosphorylation sites (GFP::Cdc27P304A,P456A) were examined, the fusion protein was found no longer to localise to mitotic chromosomes (Fig. 3C, white arrows). Thus phosphorylation on both of the two potential Cdk1 phosphorylation sites in the Drosophila Cdc27 protein is required for its chromosomal localisation. Additionally, it is noticeable that the double mutant GFP::Cdc27 protein associated with the nuclear envelope membrane less abundantly than both wild-type GFP::Cdc27 and GFP::Cdc16. Otherwise the mutant protein shows a striking similarity in its localisation patterns during the cell cycle to GFP::Cdc16 (Fig. 3C,D, white open arrows, arrowheads and double arrowheads) (Huang and Raff, 2002). It is notable that Cdc16 has no identifiable potential Cdk1 phosphorylation sites.
Fig. 3.
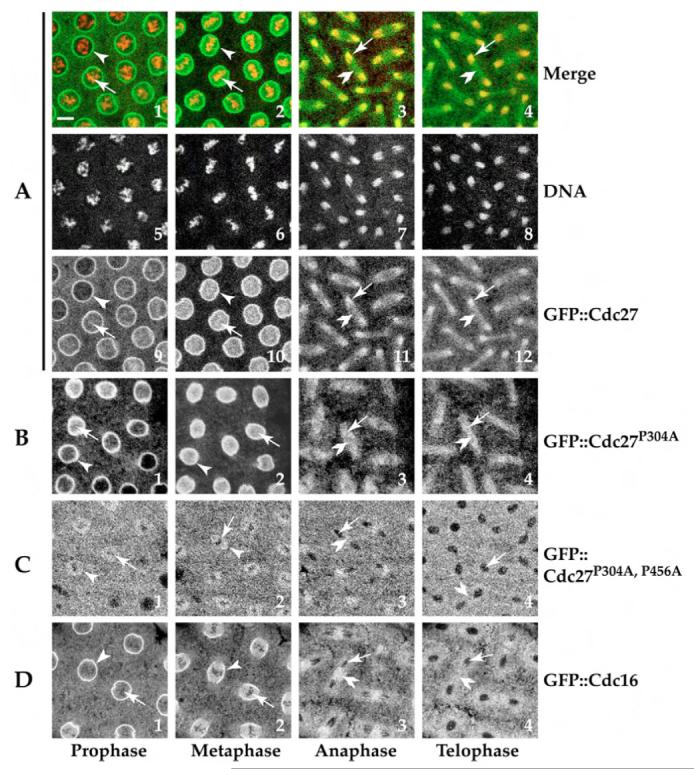
Both CDK1 phosphorylation sites are required for localisation of Cdc27 to mitotic chromosomes. (A,B) Confocal images from GFP::Cdc27P304A transgenic syncytial embryos at various stages of mitosis (B) showed no significant differences in dynamic localisation to the wild-type GFP::Cdc27 transgenic line (A). An identical result was seen with the GFP::Cdc27P456A single point mutant (not shown). However the fusion protein containing mutations in both potential Cdk1 phosphorylation sites (GFP::Cdc27P304A,P456A) was no longer found localised to the chromosomes during mitosis (C). White open arrowheads indicate the nuclear envelope membrane localised with fusion proteins (bright ring structures) in A,B,D in prophase and metaphase images. It is less abundant in C, white open arrows indicate the mitotic chromosomes regions; GFP fusion proteins in white associate with chromosomes in A and B; and do not associate with chromosomes (shadow regions) in C,D. Double white open arrowheads show the mid-body regions at anaphase and telophase. A, panels1-4, merged confocal images (GFP::Cdc27 in green, Rhodamine-H1-labelled chromosomes in red); A, panels 5-8: Rhodamine-H1-labelled chromosomes in white. Bar, 10 μm.
Cdc27 phosphorylation by Cdk1 is essential for APC/C functions
To test whether the GFP::Cdc27P304A,P456A double mutant fusion protein was still functional as an APC/C component, we introduced a copy of the gfp::cdc27P304A,P456A gene onto the second chromosome of a line that carries a mutation in the cdc27 gene (cdc27L7123) on the third chromosome. The mutant line cdc27L7123 contains a P-element insertion at 519 bp upstream of the initiating ATG of the cdc27 gene. This mutation is semi-lethal with occasional homozygous flies surviving to adulthood. These rare homozygous flies had many eye and bristle defects, were invariably sterile and only lived for a few days (http://flybase.bio.indiana.edu/.bin/fbidq.html?FBal0087027#FBrf0111595) (Deak et al., 2003; Huang and Raff, 2002). Western blot analysis of the samples made from embryos or third instar larvae brains of these flies confirmed that they expressed the GFP-Cdc27 fusion protein to a level similar to that of the endogenous Cdc27 found in wild-type embryos, but had severely reduced (<11%) levels of the endogenous Cdc27 protein (Fig. 4A-C). The cdc27L7123 (P160168) mutant stock was obtained from the Bloomington stock center. In this stock, the cdc27L7123 gene on the third chromosome is balanced using the TM3 Sb balancer chromosome. To identify and isolate homozygous cdc27L7123 larvae at different stages during development to adulthood, we replaced the TM3 Sb balancer chromosome with the TM6B Tb balancer chromosome. This balancer chromosome induces a tubby phenotype; progeny homozygous for cdc27L7123 lack the tubby phenotype. The majority of the over 200 homozygous non-tubby larvae that we isolated with an inferred genotype of gfp::cdc27 P304A,P456A/gfp::cdc27 P304A,P456; cdc27L7123/cdc27L7123, survive only to late pupation stage, as is observed in the original mutant homozygous (cdc27L7123/cdc27L7123) larvae (data not shown). Consistent with previous reports, homozygous flies hatched only rarely and these flies had many eye and bristle defects, were invariably sterile and only lived for a few days (data not shown). This finding indicates that the ectopically expressed GFP::Cdc27P304A,P456A double mutant fusion protein cannot rescue the phenotypes associated with the Cdc27L7123 mutation. Failure to rescue the mutant phenotype is not simply due to damage caused by transgene insertion at a particular locus: firstly, we found that the original transgenic GFP::Cdc27P304A,P456A homozygous line in a wild-type genetic background is completely viable and secondly, that another independently generated chromosome II insertion of gfp::cdc27P304A,P456A also failed to rescue the cdc27L7123 phenotype (data not shown). Embryonic development in cdc27L7123/cdc27L7123 and gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456; cdc27L7123/cdc27L7123 embryos are probably supported by maternally inherited endogenous wild-type Cdc27 until it is exhausted. Nevertheless, cyclin A and cyclin B protein had accumulated to high levels in the third instar larvae brains from those homozygous mutants compared with a wild-type control (Fig. 4B). This finding suggests that the APC/C function in these embryos has been disrupted. Thus it appears that phosphorylation at the two putative Cdk1 phosphorylation sites is required not only for correct protein localisation but also for the correct functioning of the APC/C that is required for normal embryonic development.
Fig. 4.
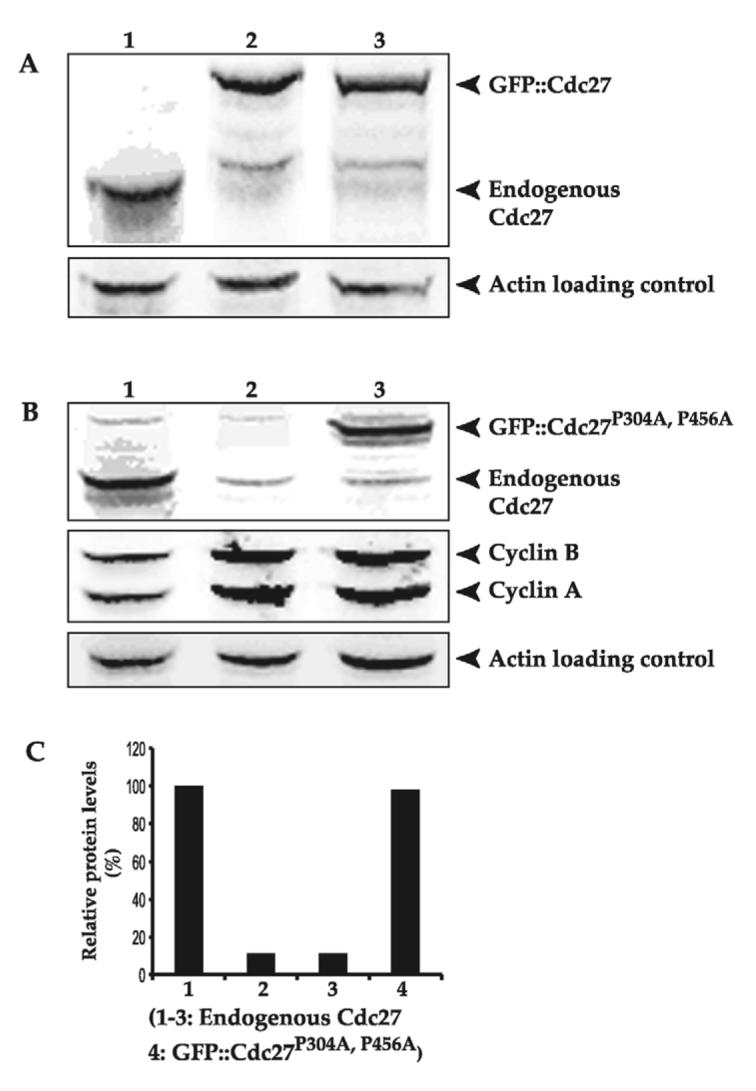
GFP::Cdc27 and GFP::Cdc27P304A can support healthy embryonic development in established stable p340 (GFP::Cdc27) and p341 (GFP::Cdc27P304A) transgenic cdc27L7123 rescue lines, but that the GFP::Cdc27P304A,P456A double mutant fusion protein does not. (A) The endogenous Cdc27 protein or GFP::Cdc27 and GFP::Cdc27P304A fusion proteins are indicated by arrows in lanes 1-3 respectively. Some (∼11%) of endogenous Cdc27 is still present in lanes 2 and 3 with the embryo samples collected from parental fly genotypes of gfp::cdc27/gfp::cdc27; cdc27L7123/cdc27L7123 and gfp::cdc27P304A/gfp::cdc27P304A; cdc27L7123/cdc27L7123 respectively. These flies carry a mutation in the cdc27gene on the third chromosome. The mutation is caused by a P-element insertion at 519 bp upstream of the ATG start site of cdc27 (http://flybase.bio.indiana.edu/.bin/fbidq.html?FBal0087027#FBrf0111595) (Huang and Raff, 2002). Actin was detected as a loading control. (B) Western blot of late third instar larvae brain samples: lane 1, W67 control; lane 2, Cdc27L7123:cdc27L7123/cdc27L7123 homozygous original mutant derived from the cdc27L7123/TM6B Tb mutant fly line; lane 3, GFP::Cdc27P304A,P456A the attempted rescue homozygous mutant (gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A; cdc27L7123/cdc27L7123) derived from the cross gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A; cdc27L7123/TM6B Tb. Endogenous Cdc27 or GFP::Cdc27 was revealed by probing with anti-Drosophila Cdc27 antibody (top panel); cyclin A and cyclin B levels are shown in the middle panel using anti-Drosophila cyclin A or cyclin B antibodies respectively; Actin (bottom panel) was detected as a loading control. (C) Relative expression levels of endogenous Cdc27 or GFP::Cdc27 in third instar larvae brains from wild-type control samples (W67), Cdc27L7123 (cdc27L7123/cdc27L7123 homozygous mutant) and GFP::Cdc27P304A,P456A (the attempted rescue homozygous mutant: gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A; cdc27L7123/cdc27L7123) were quantified from the western blot results present in the top panel of B in lanes 1-3 of the endogenous Cdc27 bands and the GFP:: Cdc27P304A,P456A band. The endogenous Cdc27 band intensity in wild-type sample is set to 100% as the comparison.
Discussion
Colchicine treatment activates the spindle checkpoint leading to high levels of both Cdk1 and Plk activity (Campbell et al., 1995; van Vugt et al., 2001; Weinert, 1997). We report here that colchicine treatment enhanced association of GFP::Cdc27 with chromosomes in syncytial embryos (Fig. 1A, panels 3 and 9, white arrows), but that there are no apparent changes in GFP::Cdc16 localisation in response to colchicine treatment (Fig. 1B, panels 3 and 9, white arrows). These data suggest that Cdk1 or Plk activity may determine the chromosomal localisation of Cdc27, in agreement with findings that Cdc27, along with several other APC subunits, are hyperphosphorylated by mitotic kinases such as Cdk1 and Plk in Xenopus, clam and human and in both fission and budding yeasts (Golan et al., 2002; Patra and Dunphy, 1998; Peters et al., 1996; Rudner and Murray, 2000; Shteinberg et al., 1999; Topper et al., 2002; Yamada et al., 1997). Cdk1/cyclin B kinase activity has been implicated in the activation of the APC/C in frog and clam (Felix et al., 1990; Shteinberg et al., 1999). Plk can be seen on centromeres and chromosomes in a variety of organisms during meiosis and mitosis (Charles et al., 1998; Glover et al., 1998; Pahlavan et al., 2000) and binds to and phosphorylates Cdc27 and some other APC/C components in human and yeast (Golan et al., 2002; Rudner and Murray, 2000). Moreover, these two kinases can be co-fractionated with isolated chromosomes in HeLa cells (Topper et al., 2002). The finding that Drosophila Cdc27 carries two putative Cdk1 phosphorylation consensus sites - sites that are absent in Cdc16 - may explain why the two core APC/C components, Cdc27 and Cdc16, respond differently to colchicine treatment. Live cell imaging taken from transgenic Drosophila syncytial embryos that ectopically express GFP::Cdc27P304A,P456A fusion protein showed that it does not localise to the mitotic chromosomes (Fig. 3C, white arrows). However, single mutations of either Cdk1 phosphorylation site (P304A or P456A) led to a localisation of the transgenically expressed fusion protein that was indistinguishable from its wild-type counterpart and functionally rescued cdc27L7123/cdc27L7123 flies (Fig. 3A,B and Fig. 4A). Therefore, it is clear that phosphorylation on both Cdk1 sites is required for proper localisation of Cdc27 to mitotic chromosomes and for normal function. However, this finding apparently contradicts a report in HeLa cells that the chromosome-associated Cdc27 fraction was not phosphorylated to the same extent as the bulk of the cellular Cdc27 (Topper et al., 2002). This apparent contradiction can be resolved in one of two ways. It can be argued that phosphorylation by Cdk1 is essential for the chromosomal localisation of Cdc27, but that once localised to the chromosome, Cdc27 is then dephosphorylated. Alternatively, it is known that a variety of kinases and phosphatases other than Cdk1/cyclin B co-fractionate with chromosomes, including Plk, Protein Phosphatase 1 and Bub3 (Topper et al., 2002); in budding yeast, it is known that the APC/C subunits Cdc27 (Apc3), Cdc16 (Apc6) and Cdc23 (Apc8) can still be phosphorylated by Cdc5 (Plk) in vitro after mutation of the phosphorylation sites targeted by Cdc28 (Cdk1) (Rudner and Murray, 2000). Thus the overall phosphorylation state of Cdc27 protein that co-fractionates with chromosomes in HeLa cells is probably determined by the multiple actions of several kinases and phosphatases: phosphorylation of the two Cdk1 sites identified in this study may well be accompanied by overall dephosphorylation of Cdc27.
The doubly mutated form of the GFP::Cdc27 fusion protein containing mutations in two putative Cdk1 phosphorylation sites is not only unable to be phosphorylated by Cdk1 kinase in vitro (Fig. 2C) and cannot associate with mitotic chromosomes in living embryos (Fig. 3C), but it also failed to rescue a cdc27 mutation (cdc27L7123), in contrast to wild-type or singly mutated fusion proteins. Homozygous third instar larvae brains collected from this attempted rescue show elevated levels of both cyclin B and cyclin A (Fig. 4B, lane 3). These observations indicate that mutation of both putative Cdk1 phosphorylation sites on Cdc27 leads to a failure of the APC/C to degrade its cyclin B and cyclin A substrates properly. The maternal contribution of endogenous wild-type Cdc27 probably supports early embryonic development because ∼11% of the endogenous Cdc27 still presents in the rescued embryos or mutant third instar larvae brain samples (Fig. 4A-C). Our results clearly show that the phosphorylation of Cdc27 by Cdk1 kinase activity is required for its mitotic chromosomal localisation and these phosphorylations are also required for its proper APC/C functions, as has been reported in other systems (Hardwick et al., 1996; Rudner et al., 2000; Rudner and Murray, 2000). It would be interesting to further distinguish the function of the chromosomal Cdc27 and we are currently addressing this problem. It is also notable that the requirement of Cdk1 phosphorylation for localisation of Cdc27 to mitotic chromosomes can explain why Cdc16, lacking Cdk1 phosphorylation sites, shows no chromosomal localisation during mitosis. It is interesting to note that the doubly mutated fusion protein associates with nuclear envelope membrane much less abundantly than wild-type protein (Fig. 3A,C, white arrows).
The absence of a signal from the doubly mutated GFP::Cdc27 on the mitotic chromosomes and the nuclear envelope cannot be due to masking by a high cytoplasmic background signals as this doubly mutated fusion protein was expressed to a similar level as wild-type GFP::CDC27 and showed a very similar level of cytoplasmic background fluorescence (Fig. 4A lane 2 and 3). In addition, the fluorescence intensities in regions other than mitotic chromosomes or nuclear envelope membrane (e.g. the mitotic spindle) are comparable with the wild-type GFP::Cdc27 confocal images in Fig. 3A,C. It is worth noting that we performed gel filtration experiments in a previous study (Huang and Raff, 2002) to show that the majority of the wild-type GFP::Cdc27 fusion protein was incorporated into the APC/C complex and it was functional. The differential localisation of APC/C components has been reported in other systems; for instance, Apc1/Tsg24 in CHO cells (Jorgensen et al., 1998) and Cdc27 (Apc3) in HeLa cells (Acquaviva et al., 2004) are centromere localised. Both Apc1/Tsg24 and Apc3 can be detected bound to isolated mitotic chromosomes, but Cdc16 has not been detected in these assays (Jorgensen et al., 1998). However, we did not formally exclude the possibility that the differential localisation of Cdc27::GFP is due to the small fraction of Cdc27::GFP protein that is not associated with the APC/C. In any event, our data indicate that localisation of Cdc27 to the nuclear membrane is directly dependent on Cdk1 kinase activity. By contrast, the localisation of Cdc16 to the nuclear envelope is not directly dependent on Cdk1 as it lacks the consensus phosphorylation sites. The interpretation of this intriguing difference will probably be that the Cdk1-kinase-dependent nuclear membrane localisation of Cdc27 is required for the recruitment of other APC/C components, including Cdc16, on to the nuclear envelope. It will be interesting to pursue how APC/C components assemble on and associate with the nuclear envelope membrane in the future.
Materials and Methods
Generation of transgenic flies
Several transgenic lines that expressed either wild-type GFP::Cdc16 or GFP::Cdc27 fusion proteins were generated by standard P-element-mediated transformation (Huang and Raff, 2002). Full-length cdc27 cDNA was modified by site-directed mutagenesis using PCR to generate constructs that contained either singly mutated putative Cdk1 phosphorylation sites or both (gfp::cdc27P304A, gfp::cdc27P456A or gfp::cdc27P304A,P456A) (Fig. 2A). The N-terminus of these modified cdc27 cDNA constructs were further modified by PCR to include an appropriate restriction enzyme site just after the initiating ATG. mGFP6 (Schuldt et al., 1998) was then subcloned in-frame into this site, and the modified gfp::cdc27 cDNAs were then subcloned into the pWR-Pubq transformation vector (Nick Brown, Wellcome/Cancer Research UK Institute, Cambridge, UK, personal communication; full cloning details available on request), which placed these cDNAs under the control of a polyubiquitin promoter. These constructs were used to generate several stably-transformed lines using standard P-element-mediated transformation methods (Robertson et al., 1988).
Rescue of the cdc27 mutation
The cdc27L7123 (P160168) mutant stock was obtained from the Bloomington stock center. The mutation is on the third chromosome and comprises a P-element insertion at 519 bp upstream of the ATG initiation codon of the cdc27 gene. The mutated gene on the third chromosome was originally balanced with the TM3 Sb balancer chromosome (http://flybase.bio.indiana.edu/.bin/fbidq.html?FBal0087027#FBrf0111595) (Deak et al., 2003; Huang and Raff, 2002). Standard genetic methods were used to generate a stock containing this cdc27L7123 mutation together with a copy of the gfp::cdc27 or its mutationally variant transgenes (gfp::cdc27P304A, gfp::cdc27P456A or gfp::cdc27P304A,P456A) on the second chromosome, and the TM3 Sb balancer chromosome was replaced with the TM6B Tb balancer chromosome.
To examine the functions of the APC/C in third instar larvae brains of the genotypes gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A, cdc27L7123/cdc27L7123 or cdc27L7123/cdc27L7123, parental flies gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A, cdc27L7123/TM6B Tb or cdc27L7123/TM6B Tb were left to lay eggs in normal fly food vials and allowed to develop to larvae. Non-Tubby larvae were then identified by their normal early morphology and isolated under a light microscope using a soft paintbrush. These homozygous larvae (genotype: gfp::cdc27P304A,P456A/gfp::cdc27P304A,P456A, cdc27L7123/cdc27L7123 or cdc27L7123/cdc27L7123) as well as W67 control larvae were then transferred to an agar plate with yeast supplied to allow further development at 22°C respectively. Third instar larvae brains from these larvae were dissected in Drosophila Ringer’s buffer and boiled in 1× SDS-PAGE loading buffer and used for western blot analysis.
Time-lapse confocal microscopy
Transgenic embryos of the appropriate genotype were observed using time-lapse confocal microscopy as described previously (Huang and Raff, 1999). A Leica TCS SP2 confocal was used for imaging. Images were transported into Adobe Photoshop or MetaMorph offline, for adjusting or quantifying the fluorescence intensities. Movies of embryos were then compiled in MetaMorph.
Immunoprecipitation and western blotting
Immunoprecipitations, SDS-PAGE, western blotting and the quantification of western blots were performed as described previously (Huang and Raff, 1999). Immunoprecipitations were carried out using high-speed extracts made from embryos expressing either the wild-type GFP::Cdc27 or mutated GFP::Cdc27 fusion proteins.
Antibodies and antibody conjugation with Dynabeads
Our own affinity-purified rabbit anti-Drosophila Cdc27, cyclin A and cyclin B polyclonal antibodies, described previously (Huang and Raff, 1999; Huang and Raff, 2002; Lim et al., 1998), were used in this study, at a 1:500 dilution. Rabbit anti-GFP polyclonal antibody was used for conjugating with Dynabeads (Invitrogen) according to the commercially available standard protocol. Mouse monoclonal [AC-15] to β-actin (Abcam) was used as a loading control.
Histone-Rhodamine conjugation
Rhodamine TRITC (Tetramethyl-rhodamine-5-isothiocyanate, λAbs 555 nm, λEm 550 nm was from Invitrogen Molecular Probes, Cat no. T1480) was conjugated with histone H1 (Calbiochem, cat no. 382150) following the protocol described (Balicki et al., 2002). The conjugated histone-Rhodamine protein was about 30-40 mg/ml in 1× injection buffer (0.1 mM Na2 HPO4, 5 mM KCl, pH 7.8) and was stored at −80°C until use. To visualise chromosomes in living embryos, about 0.2% embryo volume of this histone-Rhodamine protein in injection buffer was introduced into an appropriate stage embryo by microinjection. In this study, histone H1-Rhodamine was only used to stain chromosomes when colchicine treatment experiments were performed. It was not used when confocal time-lapse images were collected from the different GFP::Cdc27 transgenic embryos to avoid perturbing the embryonic cell cycle (unless stated otherwise).
Colchicine treatment
Colchicine was used at a final intracellular concentration of about 2.5 μM and was introduced by microinjection (stock solution 250 mM in injection buffer, about 0.1% embryo volume per injection) after several minutes histone H1-Rhodamine protein was injected (for unknown reasons colchicine can not be co-injected with histone H1-Rhodamine protein: it leads to quenching of GFP signals as well as the absence of a clearly visible histone-Rhodamine-labelled chromosome signal). Embryos are approximately 470×160 μm but can vary in length and diameter considerably, therefore, the volume of an embryo is about 6.5 nl, derived from calculation of the above dimensions after considering the volume of an ellipsoid (Parry et al., 2005). Images were normally taken for 20-30 minutes after injection to encompass at least one full nuclear division cycle.
Site-directed mutagenesis
There are two Cdk1 consensus phosphorylation sequence motifs in the Drosophila Cdc27 amino acid sequence, but there are none in Drosophila Cdc16, as determined using the Scansite, Motif Scan search engine developed by the Massachusetts Institute of Technology (http://scansite.mit.edu/). The two sequence motifs identified were T-303 SSGTPFR and S-456 QPRSPPR. Site-directed mutagenesis was performed to create the gfp::cdc27P304A, gfp::cdc27P456A or gfp::cdc27P304A,P456A mutations using the QuikChange site-directed mutagenesis kit (Stratagene). The primer pairs used for P304A were: 5′ forward: TAC GAC ATG AGC AGC GGT G(A)CT G(C)CG TTT CGC AAA CAG TTC and 3′ reverse: GAA CTG TTT GCG AAA CGC(G) AGG(T) ACC GCT GCT CAT GTC GTA; the primer pairs used for P456A were: 5′ forward: TTC GTG CAG CCA CGT G(T)CG G(C)CG CCC CGC AAA GCC AAG and 3′ reverse: CTT GGC TTT GCG GGG CGC(G) CGT(A) ACG TGG CTG CAC GAA. Transgenic flies that carry these mutated constructs then were generated using standard P-element-mediated transformation techniques (Huang and Raff, 2002).
Kinase assay
Transgenic Drosophila embryos expressing GFP, GFP::CDC27 or GFP::CDC27P304A, P456A were collected for 5 hours, the eggs were then washed and dechorionated as before (Huang and Raff, 1999). About 0.03 g of freshly dechorionated or liquid nitrogen frozen eggs was then homogenised in 100 μl of RIPA-SDS (100 mM Na Cl, 10 mM Tris-HCl pH 7.5, 0.2% SDS plus protease inhibitors: 10 μg/ml aprotinin, 10 μg/ml leupeptin, 100 μM PMSF and 1 mM benzamidine) on ice. High-speed supernatants were used for immunoprecipitation with a rabbit GFP polyclonal antibody conjugated Dynabeads (Invitrogen) as described previously. The beads after co-immuno-precipitation were washed three times in RIPA-SDS (plus protease inhibitors and phosphatase inhibitors, 50 mM Na F, B glycophosphate 10 mM and 0.1 mM NaVO4) before washing five times in Kinase wash buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM EGTA plus protease and phosphatase inhibitors). The beads were then resuspended in 40 μl of Kinase reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT plus inhibitors). Half of the immunoprecipitation was then removed for a loading control gel. After calibration, an appropriate amount of each sample was resuspended in 10 μl of Kinase reaction buffer containing 100 μM ATP, 5 μCi of [γ-32P]ATP (Amersham) and 50 ng of CDK1/cyclin B (Millipore) and placed at 30°C for 30 minutes. The reaction was terminated by the addition of 10 μl of 2× SDS-sample buffer and heating at 95°C for 5 minutes. 5 μg, 0.5 μg and 0.05 μg of histone H1 (Calbiochem) were used respectively to carry out the same reactions as a positive control. The samples were then run on a 10% SDS-PAGE precast gel (Invitrogen). The gel was washed overnight in 5% TCA, 1% sodium pyrophosphate before drying onto gel drying film (Promega) and exposing to X-ray film or signals were detected using a Phospho-Image plate on a BAS-1500 Phospho-Imager (Fuji).
Acknowledgments
This work was supported by grants to Jun-Yong Huang and Michael Whitaker from the Wellcome Trust. We thank Keith Jones for his critical reading of the manuscript, Maureen Sinclair for some technical support.
References
- Acquaviva C, Herzog F, Kraft C, Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- Balicki D, Putnam CD, Scaria PV, Beutler E. Structure and function correlation in histone H2A peptide-mediated gene transfer. Proc. Natl. Acad. Sci. USA. 2002;99:7467–7471. doi: 10.1073/pnas.102168299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer M, Braus GH, Irniger S. Two different modes of cyclin clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000;468:142–148. doi: 10.1016/s0014-5793(00)01208-4. [DOI] [PubMed] [Google Scholar]
- Campbell SD, Sprenger F, Edgar BA, O’Farrell PH. Drosophila Wee1 kinase rescues fission yeast from mitotic catastrophe and phosphorylates Drosophila Cdc2 in vitro. Mol. Biol. Cell. 1995;6:1333–1347. doi: 10.1091/mbc.6.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Deak P, Donaldson M, Glover DM. Mutations in makos, a Drosophila gene encoding the Cdc27 subunit of the anaphase promoting complex, enhance centrosomal defects in polo and are suppressed by mutations in twins/aar, which encodes a regulatory subunit of PP2A. J. Cell Sci. 2003;116:4147–4158. doi: 10.1242/jcs.00722. [DOI] [PubMed] [Google Scholar]
- Felix MA, Labbe JC, Doree M, Hunt T, Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990;346:379–382. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- Glover DM, Hagan IM, Tavares AA. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- Golan A, Yudkovsky Y, Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 2002;277:15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Raff JW. The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J. Cell Sci. 2002;115:2847–2856. doi: 10.1242/jcs.115.14.2847. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jorgensen PM, Brundell E, Starborg M, Hoog C. A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol. Cell. Biol. 1998;18:468–476. doi: 10.1128/mcb.18.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew J, Beaudette K, Litwin CM, Wang JH. Purification and characterization of a novel proline-directed protein kinase from bovine brain. J. Biol. Chem. 1992;267:13383–13390. [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- May KM, Reynolds N, Cullen CF, Yanagida M, Ohkura H. Polo boxes and Cut23 (Apc8) mediate an interaction between polo kinase and the anaphase-promoting complex for fission yeast mitosis. J. Cell Biol. 2002;156:23–28. doi: 10.1083/jcb.200106150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mirabito PM, Morris NR. BIMA, a TPR-containing protein required for mitosis, localizes to the spindle pole body in Aspergillus nidulans. J. Cell Biol. 1993;120:959–968. doi: 10.1083/jcb.120.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. The substrates of the cdc2 kinase. Semin. Cell Biol. 1991;2:261–270. [PubMed] [Google Scholar]
- Pahlavan G, Polanski Z, Kalab P, Golsteyn R, Nigg EA, Maro B. Characterization of polo-like kinase 1 during meiotic maturation of the mouse oocyte. Dev. Biol. 2000;220:392–400. doi: 10.1006/dbio.2000.9656. [DOI] [PubMed] [Google Scholar]
- Parry H, McDougall A, Whitaker M. Microdomains bounded by endoplasmic reticulum segregate cell cycle calcium transients in syncytial Drosophila embryos. J. Cell Biol. 2005;171:47–59. doi: 10.1083/jcb.200503139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev. 1998;12:2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, King RW, Hoog C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- Raff JW, Jeffers K, Huang JY. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 2002;157:1139–1149. doi: 10.1083/jcb.200203035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Hardwick KG, Murray AW. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 2000;149:1361–1376. doi: 10.1083/jcb.149.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah OJ, Ghosh S, Hunter T. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by Cdc2. J. Biol. Chem. 2003;278:16433–16442. doi: 10.1074/jbc.M300435200. [DOI] [PubMed] [Google Scholar]
- Shetty KT, Link WT, Pant HC. cdc2-like kinase from rat spinal cord specifically phosphorylates KSPXK motifs in neurofilament proteins: isolation and characterization. Proc. Natl. Acad. Sci. USA. 1993;90:6844–6848. doi: 10.1073/pnas.90.14.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 1999;260:193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper LM, Campbell MS, Tugendreich S, Daum JR, Burke DJ, Hieter P, Gorbsky GJ. The dephosphorylated form of the anaphase-promoting complex protein Cdc27/Apc3 concentrates on kinetochores and chromosome arms in mitosis. Cell Cycle. 2002;1:282–292. [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- van Vugt MA, Smits VA, Klompmaker R, Medema RH. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J. Biol. Chem. 2001;276:41656–41660. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Weinert T. A DNA damage checkpoint meets the cell cycle engine. Science. 1997;277:1450–1451. doi: 10.1126/science.277.5331.1450. [DOI] [PubMed] [Google Scholar]
- Yamada H, Kumada K, Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J. Cell Sci. 1997;110:1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]


