Abstract
Study Objective:
To assess the performance of automatic sleep scoring software (ASEEGA) based on a single EEG channel comparatively with manual scoring (2 experts) of conventional full polysomnograms.
Design:
Polysomnograms from 15 healthy individuals were scored by 2 independent experts using conventional R&K rules. The results were compared to those of ASEEGA scoring on an epoch-by-epoch basis.
Setting:
Sleep laboratory in the physiology department of a teaching hospital.
Participants:
Fifteen healthy volunteers.
Measurements and Results:
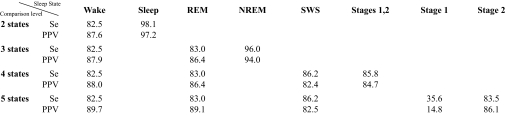
The epoch-by-epoch comparison was based on classifying into 2 states (wake/sleep), 3 states (wake/REM/NREM), 4 states (wake/REM/stages 1-2/SWS), or 5 states (wake/REM/stage 1/stage 2/SWS). The obtained overall agreements, as quantified by the kappa coefficient, were 0.82, 0.81, 0.75, and 0.72, respectively. Furthermore, obtained agreements between ASEEGA and the expert consensual scoring were 96.0%, 92.1%, 84.9%, and 82.9%, respectively. Finally, when classifying into 5 states, the sensitivity and positive predictive value of ASEEGA regarding wakefulness were 82.5% and 89.7%, respectively. Similarly, sensitivity and positive predictive value regarding REM state were 83.0% and 89.1%.
Conclusions:
Our results establish the face validity and convergent validity of ASEEGA for single-channel sleep analysis in healthy individuals. ASEEGA appears as a good candidate for diagnostic aid and automatic ambulant scoring.
Citation:
Berthomier C; Drouot X; Herman-Stoïca M; Berthomier P; Prado J; Bokar-Thire D; Benoit O; Mattout J; d'Ortho MP. Automatic analysis of single-channel sleep EEG: validation in healthy individuals. SLEEP 2007;30(11):1587-1595.
Keywords: Automatic sleep scoring, single channel, EEG, clinical validation, healthy subjects
INTRODUCTION
CONVENTIONAL POLYSOMNOGRAPHY CONSISTS IN OBTAINING RECORDINGS FROM AT LEAST 2 ELECTROENCEPHALOGRAM (EEG) CHANNELS, AN electrooculogram (EOG), and an electromyogram (EMG), and scoring them manually using Rechtschaffen and Kales rules.1 Although this method has become the reference standard, it has serious limitations. First, the recording equipment is bulky, making “real life” ambulatory recording difficult. Second, setting up the device and scoring the recordings is a time-consuming and therefore costly task. This makes conventional polysomnography unsuitable for screening large populations, for instance to investigate the impact of noise on sleep quality. In addition, the time-consuming nature of polysomnography is an obstacle to meeting the rising demand, for example, for sleep apnea diagnosis, related to its relatively high prevalence and to its cardiovascular consequences. Several automated or semi-automated methods for faster analysis of sleep recordings have been suggested. Most of them, however, require a full set of EEG, EOG, and EMG recordings.2–14
Benoit and Prado, in contrast, developed a semi-automatic analysis method based on a single EEG channel.15,16 This method was validated in both healthy volunteers and patients.17–19 A combination of multiple signal-processing techniques was then used to convert the method to a fully automated analysis of a single EEG channel recording. The analysis software is called Automatic Sleep EEG Analysis (ASEEGA, Physip, Paris, France). Following an early pilot work,20 the present study aimed at comparing sleep scoring by ASEEGA with conventional manual scoring, in healthy individuals. We used both our own data and publicly available data.
METHODS
In this section, we describe our evaluation of ASEEGA in healthy subjects. To further assess the performance and robustness of ASEEGA, we also applied it to publicly available data (see Appendix for details and results).
Study Participants and Recordings
Fifteen volunteers (aged 29.2 ± 8 years, 9 women) were enrolled in the study through local advertising. They were administered the Epworth sleepiness questionnaire and interviewed about their medical history, sleep quality and daytime sleepiness. Subjects had to be devoid of any significant medical event, to have a good sleep quality and a normal Epworth sleepiness score (<11), and to present no night symptoms. The study was approved by the local ethic committee, and volunteers gave their informed consent. In each participant, we recorded a full-night polysomnogram comprising 2 EEG channels (see below), a chin EMG, 2 EOG channels, EMGs from electrodes on the right and left tibialis anterior muscles, and an electrocardiogram (ECG), using a commercially available recording device (Embla N7000, Embla, Denver, CO, USA).
EEG activity was recorded (16 bits, 200 Hz) through 2 bipolar channels (C4-O2 and Cz-Pz, according to the international 10-20 standard system), hardware-filtered (DC, powerline and 90 Hz anti-aliasing filtering), and strongly amplified (full scale: ± 100 μV) to provide high resolution (3 nV/bit). High resolution is crucial for automatic analysis,21,22 since the signal of interest is provided by a single EEG channel.
Automatic Analysis
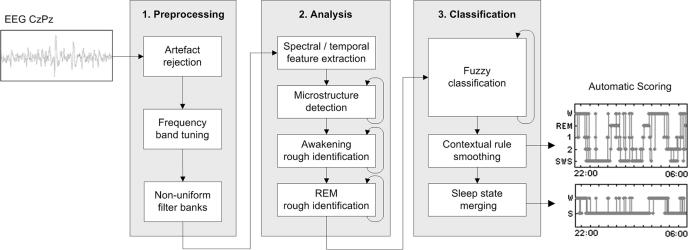
We used version R. 1.3.14 of ASEEGA. The analysis and classification algorithm has been described elsewhere.23 In brief, the automated procedure comprises 3 steps: preprocessing, analysis, and classification (see Figure 1).
Figure 1.
Diagram of the 3-step procedure for automatic sleep scoring based on a single EEG CzPz channel. The 3 steps are preprocessing, analysis, and classification.
Step 1: Preprocessing.
After downsampling of the raw signal to 100.00 Hz, artifacts are detected automatically based on signal-power criteria in specific frequency bands. Then, to accommodate the inter-individual variability of EEG signals, data-driven automated tuning of the frequency bands of interest is performed. In a given individual, the frequency band of interest can differ slightly from mean values.21,22 For instance, alpha rhythm, usually defined as 8-12 Hz activity, may occur in the 7-11 Hz range. This ability of ASEEGA to define individually tailored frequency bands should prove particularly useful when dealing with patients, whose signal tends to exhibit greater variability and instability compared to healthy individuals. In most of our healthy volunteers, the frequency bands matched conventional definitions: δ (0-4 Hz), θ (4-8 Hz), α (8-12 Hz), and σ (12-16 Hz). We used a 0-4 Hz δ band definition† rather than 0-2 Hz (the R&K definition), in accordance with Aeschbach and Borbely who showed that this frequency band (slow wave activity) better fits the description of the dynamic of slow wave sleep (SWS).24 The β band is split into β1 (16-18 Hz) and β2 (18-50 Hz) to refine the classification. Finally, the EEG signal is filtered using a nonuniform filter bank at the previously identified frequency bands.
Step 2: Analysis
The preprocessed signal is analyzed independently within each frequency band of interest. Depending on the type of EEG features to be estimated, one uses either autoregressive modelling, Fourier transform, or instantaneous frequency measurement, to extract spectral and temporal information, as well as to detect sleep microstructures (spindles, K complexes, and alpha bursts).
This analysis step also includes rough temporal localization of awakenings and REM episodes. Since resting wakefulness is usually characterized by abundant alpha frequencies and scarce delta frequencies, ASEEGA computes a contrast function defined as Cw(x)=Pα(x)Pβ1(x)/Pδ(x), where Pα(x) indicates the α power in the xth epoch. Similarly, regarding REM sleep, the preponderance of theta rhythms and high frequencies together with the low delta power is used to define the contrast function CREM(x)=Pθ(x)Pβ2(x)/Pδ(x). The maxima of the time-localization functions provide a rough estimate of the spectral signatures of wake and REM in the recording.
Step 3: Classification
Because of the EEG variability, the use of predefined sleep-stage patterns is ill-suited to automatic sleep scoring. ASEEGA uses an adaptive fuzzy logic iterative system to repeatedly update the sleep stage pattern definitions. The entire recording is then analyzed based on the final sleep-stage pattern definitions.
The first sleep stage pattern classification step consists in initializing the patterns. For each sleep stage, by combining spectral profiles of epochs that optimally satisfy scoring criteria and microstructure detection, ASEEGA defines a sleep stage pattern as a fuzzy set.‡ For example, the SWS pattern is defined by averaging the spectral composition of the epochs with the highest δ power combined with the lowest α and β powers. Stage 2 sleep is defined by averaging the spectral composition of the epochs with the highest spindle density. For the iteration (i), the ith pattern definition uses the penultimate (i-1) scoring step, considering only the epochs for which the scoring decision complies with the constraint imposed by the step 2 above. For example, an epoch scored as REM and containing a spindle cannot be selected to define the REM pattern.
Scoring is performed after each new pattern definition. Each 30-second epoch, also handled as a fuzzy set, is compared to each pattern by computing the degree of membership as the intersection of the 2 fuzzy sets. The scoring decision is based on the degree of membership and on the results of the step 2 above. Epochs that are not assignable are temporarily scored as artifacts. The final hypnogram is obtained after 3 sleep stage classification steps (initialization plus 2 repeats). Artifacts, wake episodes, and REM episodes are smoothed using contextual rules similar to visual ones. Two kinds of artifacted-epochs were distinguished:
epochs for which the EEG signal was considered as “abnormal” for a sleep EEG;
epochs to which no sleep stage was assigned.
As a consequence of the fully-automated smoothing, an artifacted-epoch between 2 Wake or REM epochs is now classified as a Wake or REM epoch. Thus accounting for temporal precedence between epochs yields a substantial decrease of the number of rejected epochs.
Manual Scoring
The 15 polysomnograms were scored by 2 sleep specialists (MH and XD) who worked independently from each other and used Rechtschaffen and Kales rules1 on 30-second epochs.
Epoch-by-Epoch Comparison
The 3 analyses were compared on an epoch-by-epoch basis. For each recording, the hypnograms were synchronized so that the onset of the first epoch was identical for the 2 scorers and for the automatic analysis. For a given epoch, 4 situations could occur: both manual scores (M1 and M2) and the automatic score (A) were identical; M1 and M2 were the same but differed from A; M1 and M2 were different; or one or both manual scorers classified the epoch as “movement artifact” and/or A classified the epoch as an artifact. For clarity and to increase the significance of the results, most of the epoch-by-epoch comparisons were based on pooled data from all 15 study participants.
Scoring Reference
The comparison between the automatic and manual sleep scoring was assessed by considering different kind of references, based on the 2 independent manual scorings mentioned above. In all comparisons, epochs scored as artifacts were discarded. First, we compared M1 to M2, A to M1, and A to M2. Then, to evaluate overall agreement among the three scorings, we performed 2 additional comparisons: an overall comparison of A, M1, and M2; and a comparison of A to M1 and M2 confined to those epochs assigned the same score by the 2 manual scorers (M1∩M2). M1∩M2 was used as the reference standard throughout the validation procedure.
Comparison Levels
A, M1, and M2 were compared at 4 different levels of sleep state discrimination: 2-state scoring (wake or sleep; with the sleep category combining stage 1, stage 2, SWS, and REM), 3-state scoring (wake/REM/NREM), 4-state scoring (wake/REM/stage 1 or 2/SWS), and 5-state scoring (wake/REM/stage 1/stage 2/SWS).
Statistics
To evaluate quantitatively the automatic scoring obtained with ASEEGA, we computed several complementary measures which correspond to the most common criteria used in the literature. They are listed and defined hereafter.
Percentage Agreement
Epoch-by-epoch agreement was defined as the percentage of epochs that were assigned the same state. Scoring agreement was determined for all pairwise comparisons and for the A versus M1∩M2 comparison.
Sensitivity and Positive Predictive Value
In comparing A and M1∩M2, we also computed the sensitivity and positive predictive value (PPV) associated with each state. As defined by Altman,25 sensitivity is the proportion of positives that are correctly identified by A, while PPV is the proportion of epochs with positive test results that are correctly diagnosed.§ These parameters complement each other. Sensitivity reflects the performance of ASEEGA compared to the current reference standard. PPV estimates the reliability of automatic scoring, should this method be used more widely, in the absence of a standard reference for comparison.**
Cohen's Kappa
Cohen's kappa (κ)26 was used to assess agreement for the pairwise comparisons and for the overall comparison of A, M1, and M2. Kappa corrects for the probability of agreement due to chance alone and quantifies agreement among 2 or more scorers. In our study, the 2 manual scorers, who were both sleep specialists working in the same unit, produced similar scores. As a result, κ values associated with the overall comparison chiefly reflect the performance of ASEEGA. In addition, we computed the global κ values for each individual study participant, which we denoted κ∗. These global values provided an assessment of the variability in ASEEGA performance across individuals.
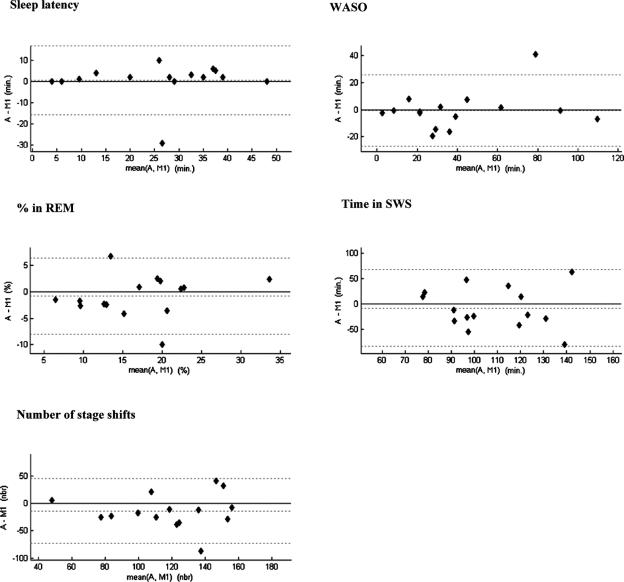
Bland and Altman Plots
The reliability of the sleep structure provided by ASEEGA was assessed qualitatively by constructing Bland and Altman plots27 for sleep latency (time from lights off to the first stage-2 epoch), wake after sleep onset (WASO), REM sleep percentage, SWS duration and number of stage shifts. M1 and M2 were similar for these 5 parameters. For clarity, only comparisons with M1 are reported here.
To summarize, the epochs for which the 2 experts disagreed were included in both the pairwise comparisons (including Bland and Altman plots, Fig. 3) and the overall comparison (global kappa), but were discarded when comparing ASEEGA to the defined reference (the expert-consensus hypnogram). These comparisons are complementary and thus prevent bias in the evaluation, either for or against our approach.
Figure 3.
Bland-Altman plots of sleep parameters showing the differences between ASEEGA scoring and manual scoring by scorer 1 versus the corresponding mean, calculated for sleep latency (min), wake after sleep onset (WASO, min), REM sleep (%), slow wave sleep (SWS, min) and number of stage shifts. The mean difference and the limits of agreement (± 1.96 SD) are represented as dotted lines. A similar figure could be drawn with the results from scorer 2.
RESULTS
According to M1, mean (± SD) sleep period time (SPT) was 456 ± 32 min and mean total sleep time was 415 ± 42 min. On average, the study participants spent 112 ± 29 min in SWS, 202 ± 46 min in stage 2, and 24 ± 8 min in stage 1. The mean proportion of SPT spent in REM sleep was 17% ± 6%. Mean WASO was 42±29 min and mean arousal index was 17.7 ± 7/h. The relationship between the scorings and the different sleep stages can be summarized in contingency tables. The most general one, obtained for the five-state classification, is reported in Table 1. All the metrics detailed below were computed from the contingency tables obtained for the different levels of classification. Moreover, as an illustration example, Figure 2 shows the details of the results from one subject who took part in this study.
Table 1.
Contingency Table
| M1 & M2 |
|||||||
|---|---|---|---|---|---|---|---|
| Artifact | Wake | REM | Stage 1 | Stage 2 | SWS | ||
| ASEEGA | Artifact | 2 | 6 | 11 | 1 | 1 | 27 |
| Wake | 213 | 1,609 | 59 | 88 | 37 | 0 | |
| REM | 286 | 52 | 1,749 | 24 | 139 | 0 | |
| Stage 1 | 211 | 136 | 105 | 85 | 250 | 0 | |
| Stage 2 | 1,141 | 134 | 191 | 41 | 4,534 | 369 | |
| SWS | 313 | 20 | 2 | 1 | 467 | 2,303 | |
Contingency table of 5-state epoch classification: automated analysis versus manual scoring (A vs. M1∩M2). Epochs with “artifacts” by ASEEGA analysis were discarded (first row). Epochs labeled as “movement” by at least one of the manual scorers together with epochs that were assigned different scores by the 2 manual scorers were discarded (first column).
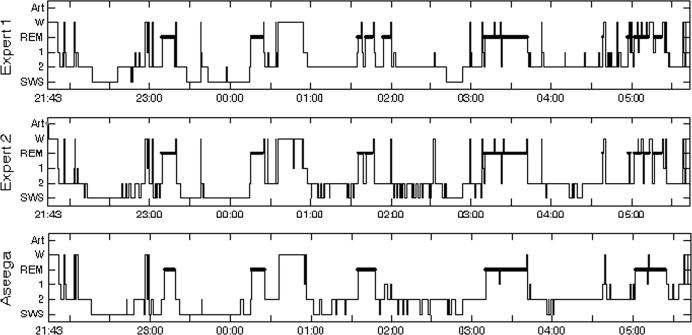
Figure 2.
Representative 5-state hypnograms obtained using ASEEGA (bottom) and manual scoring (scorer 1, top panel; and scorer 2, middle panel). For this subject, agreement between M1 and M2 was 79% (κ = 0.70), 73% between M1 and A (κ = 0.62), and 77% between M2 and A (κ = 0.66). M1 and M2 scores differed in 198 epochs. Finally, agreement between A and M1∩M2 was 83%.
Epochs Kept for the Analyses
The comparisons between scorers as well as the agreement and kappa coefficients have been computed based on the total of 14,607 epochs from the 15 study participants. Of these epochs, 48 (0.3%) were classified as artifact by ASEEGA and 27 (0.2%) were classified as “movement time” by one or both manual scorers. Some epochs were rejected for more than one reason; the total number of discarded epochs was 52, leaving 14,555 epochs for the comparisons. Increasing the comparison level from 2-state scoring to 5-state scoring resulted in a slight decrease in agreement between M1 and M2. For instance, an epoch scored as REM by M1 and as stage 1 by M2 was considered to reflect agreement between the 2 scorers in the 2-state scoring but disagreement in higher-level scorings.
Two-State Scoring: Wake/Sleep
Agreement was 97.2% (κ = 0.89) between M1 and M2, 95.2% (κ = 0.80) between M1 and A, and 94.1% (κ = 0.76) between M2 and A. For the overall comparison, κ was 0.82 (κ∗=0.79 ± 0.09, mean ± SD). M1 and M2 scores differed for 423 epochs. Agreement between A and M1∩M2 was 96.0%.
Three-State Scoring: Wake/REM/NREM
Agreement was 94.4% (κ = 0.88) between M1 and M2, 89.6% (κ = 0.77) between M1 and A, and 89.5% (κ = 0.78) between M2 and A. For the overall comparison, κ was 0.81 (κ∗ = 0.80 ± 0.06). M1 and M2 scores differed for 834 epochs. Agreement between A and M1∩M2 was 92.1%.
Four-State Scoring: Wake/REM/Stages 1 and 2/SWS
Agreement was 87.9% (κ = 0.82) between M1 and M2, 79.6% (κ = 0.70) between M1 and A, and 81.2% (κ = 0.72) between M2 and A. For the overall comparison, κ was 0.75 (κ∗ = 0.74 ± 0.06). M1 and M2 scores differed for 1779 epochs. Agreement between A and M1∩M2 was 84.9%.
Five-State Hypnogram: Wake/REM/Stage1/Stage 2/SWS
Agreement was 85.3% (κ = 0.80) between M1 and M2, 76.0% (κ = 0.67) between M1 and A, and 78.2% (κ = 0.69) between M2 and A. For the overall comparison, κ was 0.72 (κ∗ = 0.71 ± 0.07). M1 and M2 scores differed for 2160 epochs. Agreement between A and M1∩M2 was 82.9%.
Sensitivity and Positive Predictive Value
Sensitivity and PPV values for each of the 4 levels of comparison are reported in Table 2.
Table 2.
Automatic vs. Manual Scoring
Sensitivity (Se, %) and positive predictive value (PPV, %) for each wake and sleep state according to the various comparison levels (2-, 3-, 4-, and 5-state hypnograms).
Bland and Altman Plots
Figure 3 shows the Bland-Altman plots comparing A and M1 estimates of the sleep parameters. No systematic trend was found between the difference and the mean of any of the sleep parameters. In most cases, similar results were obtained when A was compared to M2 (data not shown). However, it appears that the percentage of time spent in REM is substantially different for 2 subjects out of 15. In the first case, the respective estimations are 17% for A, 10% for M1, and 15% for M2, respectively. In the second case, M1 and M2 estimations are much closer (25% and 22% respectively) while the automatic estimation using A is 15%. However, the latter recording yields the worst between-expert-agreement (67%). Our conclusion is that those 2 extreme cases that yield a substantial variability between experts might be due to a poor recorded signal and could be seen as outliers. Similarly, concerning the time spent in SWS and assuming the 2 visual scoring can be taken as repeated measurements, we computed the coefficients of repeatability (CR).27 Obtained CR was 66 min, showing that this sleep parameter is less reliable than any other. For the A-M1 comparison, 14 out of 15 points were found in the corresponding range. For the worst dataset, A=99 min and M1=179 min, but M2 found 98 min.
Finally, as shown in the last graph of Figure 3, the automatic analysis detects less stage shifts than the visual one, yielding a less fragmented hypnogram.
Quantitative Interpretation of the Cohen's Kappa
We propose here 2 ways of interpreting the obtained kappa values. First, since calculated out of a large number of observations (epochs), an asymptotic standard error can be associated with our kappa values28 and the corresponding z statistics can be computed. Whatever the comparison in our study, the z proved highly significant against the mean-centred and reduced normal law.
A more heuristic way of interpreting the Kappa values was proposed by Landis and Koch29 and is reported in Table 3. Note that according to this table, all the observed agreements appear to be good, if not excellent.
Table 3.
Level of Agreement and Kappa Values
| Agreement | Kappa |
| Excellent | > 0.8 |
| Good | 0.61 – 0.80 |
| Reasonable | 0.41 – 0.60 |
| Poor | 0.21 – 0.40 |
| Bad | 0 – 0.20 |
| Very Bad | < 0 |
Qualitative interpretation of kappa values in terms of level of agreement between scorers, as proposed by Landis and Koch.29
DISCUSSION
Our study demonstrates that fully automated single-EEG-channel analysis provides reliable 5-stage sleep scoring that exhibits 82.9% agreement with a manually scored standard reference, defined as the consensus between 2 scorers who used Rechtschaffen and Kales rules. ASEEGA was more than 82% sensitive for detecting wakefulness, sleep stage 2, slow wave sleep, and REM sleep; and the associated PPV values also exceeded 82%. ASEEGA correctly identified the vast majority of epochs, even when 5 sleep stages were distinguished. Moreover, the kappa coefficient κ associated with the pooled dataset was never below 0.72, indicating a high level of coherence between the 2 manual scorers and ASEEGA.
To assess the reliability and validity of ASEEGA, its performance must be carefully compared to those obtained by the manual scorers. Defining the standard reference is the first task. Several studies compared 5-stage manual scoring by experts working at different sleep centers. Interscorer agreements ranged from 61% to 96% for healthy individuals30,31 and from 65% to 78% for patients.30,32 Inter-laboratory agreement was significantly lower than intra-laboratory agreement. In the Sleep Heart Health Study,33 the upper bound of interscorer agreement was estimated by training the scorers in the use of the same scoring rules, in order to minimize interscorer disagreement. With recordings from patients, mean pairwise agreement among 3 scorers was 87% and mean intrascorer agreement was 88%.34 These values are closely similar to those obtained in our study with the 5-state scoring system. Importantly, the 2 scorers were neither involved in the development, nor in the tuning of the algorithm. They are sleep specialists, belonging to the same medical unit and use the same scoring technique. Their scorings were independent of each other. Therefore, interscorer agreement in our study was probably overestimated compared to the conditions of everyday practice. Such overestimation would create bias against ASEEGA. Despite this potential bias, ASEEGA proved an effective and robust tool. Note that considering only 2 independent scorers is debatable31 and here corresponds to a compromise between having more than one reference to be able to evaluate interscorer variability, on the one hand, and coping with practical constraints of having several medical experts to analyse the whole cohort hypnograms, epoch-by-epoch, on the other hand.
Of the many attempts to automate sleep staging, some studies focus on patient population or do not perform R&K hypnogram scoring,4,5,9,14,35 which prevents any direct and fair statistical comparison with our results. In this study, we focus on evaluating the R&K scoring of our original approach, on a full cohort of healthy subjects, and thus compare our results with studies that involved epoch-by-epoch comparison to manual scoring in healthy individuals. Most of these methods relied on 2 EEG channels, one EOG, and one EMG. The validation studies, which involved variable numbers of individuals and 4 to 6 sleep states, showed 69% to 90% agreement with manual scoring.2,3,6–8,10,11,13 Combining sleep stages 3 and 4 into a single SWS sleep state is common practice and yields a 5-state scoring system. We used this approach. ASEEGA performed similarly to the previously reported methods but required only a single EEG channel.
Differentiating REM from stage 1 or wakefulness by automatic sleep analysis has been reported to be difficult.21,22 REM sleep criteria in the R&K rules used for manual scoring include presence of rapid eye movements and muscle atonia.1 However, EEG spectral analysis showed a distinctive frequency combination during REM sleep, with very low delta and sigma power co-existing with greater power in higher frequencies (15-35 Hz).24,31 This combination of criteria has been suggested as a specific EEG marker for REM sleep,36–38 providing the rationale for the CREM contrast function of ASEEGA, in which the ratio between β2 (18-50 Hz) and δ powers serves to discriminate between REM sleep and other sleep stages, without EOG or EMG. Similar automated sleep analysis and REM sleep detection methods using a single EEG channel have been investigated in rats,39 and the results support the ability of complex EEG analysis to detect REM sleep without EOG recordings.
Although REM, stage 1, and wakefulness can be distinguished based on their spectral composition,40 ASEEGA uses 2 original algorithms, a feature that may explain its good performance. First, ASEEGA can identify the baseline resting EEG frequency of each individual, which may differ slightly from the usual value. For instance, a baseline frequency with a maximal power at 7 or 13 Hz is classified by ASEEGA as wakefulness, although these values are below and above the usual range, respectively. Subsequently, ASEEGA automatically adjusts the spectral criteria for spindle frequency. For instance, if the baseline resting EEG frequency peaks at 13 Hz, the frequency criteria for spindle are automatically shifted to 14-18 Hz. In this healthy subject study, if most recordings did not require any frequency band tuning (alpha main rhythms ranged from 9.5 Hz to 11.2 Hz), one recording requested a shift of +0.5 Hz (alpha main rhythm was detected at 12.0 Hz) and one requested a shift of −0.5 Hz (alpha main rhythm was detected at 8.8 Hz). The second original algorithm is a self-learning paradigm that ensures repeated optimization of the sleep stage patterns based on the data recorded up to each optimization time point. This enables the algorithm to optimize pattern specificity by relying on the epochs that contain the most clearly distinguishable sleep stages.
This study was limited to healthy volunteers and was done without a previous habituation night. The volunteers reported poor sleep conditions in the laboratory. In accordance with this subjective feeling, the number of arousals per hour of sleep was high. However, it is obviously not tenable to fully infer the algorithm performance in a real clinical setting from the current results. A full evaluation study on patients is required and will be the focus of a subsequent work.
Nevertheless, to further assess and demonstrate the abilities of ASEEGA in healthy subjects, we also applied it to a publicly available sleep database (see the Appendix for all details). The recording characteristics of the test database did not comply with the algorithm recommendations, especially in terms of recording sites (Pz-Oz instead of Cz-Pz) and analogical gain (50 times lower in the public dataset). As stated previously, these 2 parameters are of great importance, since ASEEGA is based on a single-channel signal only. However, the performance obtained on the 2 cohorts are very similar for a 2-state classification (Wake/Sleep scoring). Interestingly, only when higher levels of discrimination are required, the results obtained on the public dataset are poorer than the ones obtained on the new corpus. The latter illustrates that, the more stages one wants to discriminate, the higher the importance of the quality of the signal one feeds the algorithm with; namely in terms of amplitude resolution and recording site.
In summary, we compared the ASEEGA automated sleep analysis program based on a single EEG channel to conventional manual scoring of a full polysomnogram by sleep specialists. Sensitivity and PPV of ASEEGA for sleep stage detection were similar to those provided by automatic approaches that rely on multichannel recordings, which is very promising for future clinical applications. These results in young healthy individuals need to be extended to older healthy individuals and to patients. Applications that can be anticipated based on the different levels of analysis include field studies and large-scale population studies.
ACKNOWLEDGMENTS
We extend our warm thanks to Dr F. Goldenberg for her helpful discussions, G. Rouffet for technical support, A. Hichkat and L. Margarit for technical help, and G. De Peretti for fruitful statistical discussions.
APPENDIX
The algorithm had been tested on the Physionet public database that provides sleep recordings and corresponding hypnograms in European Data Format (http://www.physionet.org/physiobank/database/). It includes the recordings (16 bits, 100 Hz) of 8 healthy subjects (21-35 years old) with no medication. We used the Pz-Oz channel only; which was the closest recording site to ASEEGA requirements (Cz-Pz).
The overall agreements (from ~8500 epochs for the 8 recordings) between ASEEGA and the known reference were 95.4%, 88.3%, 74.5%, and 71.2%, for the 2-state to 5-state scoring, respectively. The corresponding kappa coefficients were 0.83, 0.76, 0.63, and 0.61, respectively. Finally, the sensitivity of ASEEGA to detect wakefulness, in a 5-state classification, was 85.2% (PPV = 87.5%). Similarly, the sensitivity to detect REM is 63% (PPV = 91.7%).
Footnotes
Disclosure Statement
This work was supported in part by the Centre Régional d'Innovation et de Transfert de Technologie Biomédicale (BIOCRITT). Dr. Christian Berthomier is the CEO, co-creator, shareholder, and an employee of the Physip S.A Company. Dr. Drouot has participated in research supported by Respironics and has participated in a speaking engagement for UCB Laboratories. Dr. Pierre Berthomier is co-creator, shareholder, and an employee of the Physip S.A Company. Dr. d'Ortho has participated in research supported by Respironics, Resmed, Guidant, and GlaxoSmithKline. The other authors have indicated no financial conflicts of interest.
FOOTNOTES
Note that this might cause a slight increase in the misclassification of stage 2 epochs that would then be labeled as SWS, especially in the presence of several K complexes. However, this has to be tempered when dealing with older subjects whose δ rhythm may increase in frequency. At the expense of some classification errors, our objective here is to be generic and to adopt a fair trade-off in order not to miss fast δ activity.
A fuzzy set is a collection of objects with membership values between 0 (complete exclusion) and 1 (complete membership). The membership values are continuous and express the degrees to which each object is “compatible” with the properties or features that are specific to the collection. Thus using a partial quantification of membership, fuzzy logic enables us, during the iterative classification step, to deal with epochs that could possibly belong to different sleep stages.
Sensitivity and PPV differ by their denominator only. In our context, sensitivity is the ratio between the number of well classified epochs and the total number of epochs genuinely in that same state. PPV is the ratio between the number of well classified epochs and the total number of epochs classified in that same state by our approach (A).
PPV is also the conditional probability of obtaining the correct classification by automatic scoring.
REFERENCES
- 1.Rechtschaffen A, Kales A. A manual of standardized terminology, techniques and scoring systems for sleep stages of human subjects. Washington DC: National Health Institutes; 1968. [Google Scholar]
- 2.Agarwal R, Gotman J. Computer-assisted sleep staging. IEEE Trans Biomed Eng. 2001;48:1412–23. doi: 10.1109/10.966600. [DOI] [PubMed] [Google Scholar]
- 3.Anderer P, Gruber G, Parapatics S, et al. An E-health solution for automatic sleep classification according to Rechtschaffen and Kales: validation study of the Somnolyzer 24 x 7 utilizing the Siesta database. Neuropsychobiology. 2005;51:115–33. doi: 10.1159/000085205. [DOI] [PubMed] [Google Scholar]
- 4.Caffarel J, Gibson GJ, Harrison JP, Griffiths CJ, Drinnan MJ. Comparison of manual sleep staging with automated neural network-based analysis in clinical practice. Med Biol Eng Comput. 2006;44:105–10. doi: 10.1007/s11517-005-0002-4. [DOI] [PubMed] [Google Scholar]
- 5.Fischer Y, Junge-Hulsing B, Rettinger G, Panis A. The use of an ambulatory, automatic sleep recording device (QUISI version 1.0) in the evaluation of primary snoring and obstructive sleep apnoea. Clin Otolaryngol Allied Sci. 2004;29:18–23. doi: 10.1111/j.1365-2273.2004.00759.x. [DOI] [PubMed] [Google Scholar]
- 6.Kemp B, Groneveld EW, Janssen AJ, Franzen JM. A model-based monitor of human sleep stages. Biol Cybern. 1987;57:365–78. doi: 10.1007/BF00354982. [DOI] [PubMed] [Google Scholar]
- 7.Kubicki S, Holler L, Berg I, Pastelak-Price C, Dorow R. Sleep EEG evaluation: a comparison of results obtained by visual scoring and automatic analysis with the Oxford sleep stager. Sleep. 1989;12:140–9. doi: 10.1093/sleep/12.2.140. [DOI] [PubMed] [Google Scholar]
- 8.Kuwahara H, Higashi H, Mizuki Y, Matsunari S, Tanaka M, Inanaga K. Automatic real-time analysis of human sleep stages by an interval histogram method. Electroencephalogr Clin Neurophysiol. 1988;70:220–9. doi: 10.1016/0013-4694(88)90082-x. [DOI] [PubMed] [Google Scholar]
- 9.Pittman SD, MacDonald MM, Fogel RB, et al. Assessment of automated scoring of polysomnographic recordings in a population with suspected sleep-disordered breathing. Sleep. 2004;27:1394–403. doi: 10.1093/sleep/27.7.1394. [DOI] [PubMed] [Google Scholar]
- 10.Prinz PN, Larsen LH, Moe KE, Dulberg EM, Vitiello MV. C STAGE, automated sleep scoring: development and comparison with human sleep scoring for healthy older men and women. Sleep. 1994;17:711–7. [PubMed] [Google Scholar]
- 11.Schaltenbrand N, Lengelle R, Toussaint M, et al. Sleep stage scoring using the neural network model: comparison between visual and automatic analysis in normal subjects and patients. Sleep. 1996;19:26–35. doi: 10.1093/sleep/19.1.26. [DOI] [PubMed] [Google Scholar]
- 12.Sforza E, Vandi S. Automatic Oxford-Medilog 9200 sleep staging scoring: comparison with visual analysis. J Clin Neurophysiol. 1996;13:227–33. doi: 10.1097/00004691-199605000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Stanus E, Lacroix B, Kerkhofs M, Mendlewicz J. Automated sleep scoring: a comparative reliability study of two algorithms. Electroencephalogr Clin Neurophysiol. 1987;66:448–56. doi: 10.1016/0013-4694(87)90214-8. [DOI] [PubMed] [Google Scholar]
- 14.White DP, Gibb TJ. Evaluation of a computerized polysomnographic system. Sleep. 1998;21:188–96. [PubMed] [Google Scholar]
- 15.Bouard G, Benoit O, Lacombe J. Real-time analysis and semi-automatic classification of sleep EEG signals using a microprocessor. Part II: Application to normal and disturbed sleep. In: Court L, Trocherie S, Doucet J, editors. Le traitement du signal en électrophysiologie expérimentale et clinique du système nerveux central. Candé, France: Presses Lefrancq; 1986. pp. 531–44. [Google Scholar]
- 16.Le Roux J, Moreau N, Prado J. Real-time analysis and semi-automatic classification of sleep EEG signals using a microprocessor. Part I: Principles of analysis. In: Court L, Trocherie S, Doucet J, editors. Le traitement du signal en électrophysiologie expérimentale et clinique du système nerveux central. Candé, France: Presses Lefrancq; 1986. pp. 521–30. [Google Scholar]
- 17.Benoit O, Bouard G, Payan C, Borderies P, Prado J. Effect of a single dose (10 mg) of zolpidem on visual and spectral analysis of sleep in young poor sleepers. Psychopharmacology (Berl) 1994;116:297–303. doi: 10.1007/BF02245332. [DOI] [PubMed] [Google Scholar]
- 18.Benoit O, Daurat A, Prado J. Slow (0.7-2 Hz) and fast (2-4 Hz) delta components are differently correlated to theta, alpha and beta frequency bands during NREM sleep. Clin Neurophysiol. 2000;111:2103–6. doi: 10.1016/s1388-2457(00)00470-3. [DOI] [PubMed] [Google Scholar]
- 19.Daurat A, Aguirre A, Foret J, Benoit O. Disruption of sleep recovery after 36 hours of exposure to moderately bright light. Sleep. 1997;20:352–8. [PubMed] [Google Scholar]
- 20.Berthomier C, Drouot X, Herman-Stoïca M, et al. A Wake-REM-NREM automatic classification based on a single EEG channel: epoch by epoch comparison with human sleep scoring in healthy subjects. Sleep Medicine. 2005;Vol. 6(Suppl.2):S194. [Google Scholar]
- 21.Hasan J. Past and future of computer-assisted sleep analysis and drowsiness assessment. J Clin Neurophysiol. 1996;13:295–313. doi: 10.1097/00004691-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Penzel T, Conradt R. Computer based sleep recording and analysis. Sleep Med Rev. 2000;4:131–48. doi: 10.1053/smrv.1999.0087. [DOI] [PubMed] [Google Scholar]
- 23.Berthomier C, Prado J, Benoit O. Automatic sleep EEG analysis using filter banks. Biomed Sci Instrum. 1999;35:241–6. [PubMed] [Google Scholar]
- 24.Aeschbach D, Borbely AA. All-night dynamics of the human sleep EEG. J Sleep Res. 1993;2:70–81. doi: 10.1111/j.1365-2869.1993.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 25.Altman DG. Practical statistics for medical research. London: Chapman and Hall; 1991. Diagnostic tests; pp. 409–19. [Google Scholar]
- 26.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20:37–46. [Google Scholar]
- 27.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–10. [PubMed] [Google Scholar]
- 28.Fleiss JL, Cohen J, Everitt BS. Large sample standard errors of kappa and weighted kappa. Psychol Bull. 1969;72:323–27. [Google Scholar]
- 29.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 30.Norman RG, Pal I, Stewart C, Walsleben JA, Rapoport DM. Interobserver agreement among sleep scorers from different centers in a large dataset. Sleep. 2000;23:901–8. [PubMed] [Google Scholar]
- 31.Ferri R, Ferri P, Colognola RM, Petrella MA, Musumeci SA, Bergonzi P. Comparison between the results of an automatic and a visual scoring of sleep EEG recordings. Sleep. 1989;12:354–62. [PubMed] [Google Scholar]
- 32.Danker-Hopfe H, Kunz D, Gruber G, et al. Interrater reliability between scorers from eight European sleep laboratories in subjects with different sleep disorders. J Sleep Research. 2004;13:63–9. doi: 10.1046/j.1365-2869.2003.00375.x. [DOI] [PubMed] [Google Scholar]
- 33.Quan SF, Howard BV, Iber C, et al. The Sleep Heart Health Study: design, rationale, and methods. Sleep. 1997;20:1077–85. [PubMed] [Google Scholar]
- 34.Whitney CW, Gottlieb DJ, Redline S, et al. Reliability of scoring respiratory disturbance indices and sleep staging. Sleep. 1998;21:749–57. doi: 10.1093/sleep/21.7.749. [DOI] [PubMed] [Google Scholar]
- 35.Royston R, Aldridge A. Clinical evaluation of additional single channel EEG in a home sleep studies programme. Journal of Sleep Research. 2002;Vol.11(Suppl.1):197. [Google Scholar]
- 36.Ferri R, Elia M, Musumeci SA, Pettinato S. The time course of high-frequency bands (15-45 Hz) in all-night spectral analysis of sleep EEG. Clin Neurophysiol. 2000;111:1258–65. doi: 10.1016/s1388-2457(00)00303-5. [DOI] [PubMed] [Google Scholar]
- 37.Mann K, Roschke J. Different phase relationships between EEG frequency bands during NREM and REM sleep. Sleep. 1997;20:753–6. doi: 10.1093/sleep/20.9.753. [DOI] [PubMed] [Google Scholar]
- 38.Uchida S, Maloney T, Feinberg I. Sigma (12-16 Hz) and beta (20-28 Hz) EEG discriminate NREM and REM sleep. Brain Res. 1994;659(1-2):243–8. doi: 10.1016/0006-8993(94)90886-9. [DOI] [PubMed] [Google Scholar]
- 39.Karasinski P, Stinus L, Robert C, Limoge A. Real-time sleep-wake scoring in the rat using a single EEG channel. Sleep. 1994;17:113–9. doi: 10.1093/sleep/17.2.113. [DOI] [PubMed] [Google Scholar]
- 40.Corsi-Cabrera M, Munoz-Torres Z, del Rio-Portilla Y, Guevara MA. Power and coherent oscillations distinguish REM sleep, stage 1 and wakefulness. Int J Psychophysiol. 2006;60:59–66. doi: 10.1016/j.ijpsycho.2005.05.004. [DOI] [PubMed] [Google Scholar]