Abstract
Rhabdovirus is a negative strand RNA virus that packages a ribonucleorptein (RNP) complex. The RNP is composed of a genome that is encapsidated completely by the nucleoprotein (N). Structural comparisons of the RNA-nucleoprotein complexes from two members, vesicular stomatitis virus (VSV) and rabies virus (RABV), revealed highly conserved characteristics of folding, RNA binding, and assembly despite their lack of significant homology in amino acid sequence. The RNA binding cavity is located between two conserved domains formed by α–helices, but the positively charged residues that coordinate with the phosphate groups are at different sites. The intermolecular interactions among N molecules have a conserved pattern that is rendered, however, by different residues. The curvature of the RABV N-RNA complex in the crystal structure is larger than that of the VSV N-RNA complex. The more relaxed curvature allows the bases in the RNA to stack more tightly, and at the same time, the helices near the C-terminus move into the created space in order to cover the bound RNA. This may explain how the RNP can adopt different conformations from being packed as a superhelix in the virion to a relaxed linear structure once being delivered into the cytoplasm.
Keywords: Negative strand RNA virus, ribonucleoprotein, assembly, RNA encapsidation, nucleoprotein, conserved structure
Rhabdovirus is a negative strand RNA virus belonging to the Mononegavirale order. A rhabdovirus virion is shaped like a bullet that packages a ribonucleoprotein (RNP) as a superhelix inside the viral envelope. The RNP is composed of a single strand RNA genome encapsidated completely by the viral nucleoprotein. The genome encodes five viral genes: the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large subunit of the viral RNA-dependent RNA polymerase (L). The virion enters the cell through receptor attachment and membrane fusion carried out by the G protein. After entry, the RNP is released to serve as the template for transcription and replication. The structure of the RNP is essential for virus assembly and RNA synthesis because the genomic RNA is always encapsidated by the N protein in every step of the virus replication cycle. Crystal structures of the N protein in complex with RNA have been determined for two members of the rhabdovirus family, vesicular stomatitis virus (VSV) [Green et al., 2006], and rabies virus (RABV) [Albertini et al., 2006]. Structural comparisons of the two complex structures revealed some conserved characteristics of the N protein which may provide some insights on the functions of the N protein with regard to viral assembly and replication.
The structure of both N-RNA complexes has a ring-like assembly and encapsidates a single strand RNA with random sequence. The ring of the VSV N-RNA complex contains 10 N protein molecules, and that of the RABV N-RNA complex, 11 N protein molecules. Each N protein molecule is associated with 9 nucleotides. When the structure of a single VSV N molecule (PDB code 2GIC) was superimposed with that of a single RABV N molecule (PDB code 2GTT) by use of the DALI program [Holm & Park, 2000], 376 residues are aligned between the two structures with a RMSD of 2.5 Å in Cα positions, and a Z-score of 33.4 (Figure 1). This suggests that the overall structure of the two N proteins is highly homologous even though the homology of their amino acid sequences is very low (15% identity) [Walker et al., 1994]. On the other hand, there are some interesting differences in local structural features despite of high homology in their overall structures. There are 422 amino acids in the VSV N protein and 450 amino acids in the RANV N protein, which indicates that the structure in some regions of the two proteins should be different. At the first place, residue Ser2 of the VSV N protein is aligned with residue Lys5 of the RABV N protein. The first four residues in the RABV N proteins are disordered in this crystal structure. The N-terminus of the RABV N protein was shown to be required for binding with the chaperone P protein [Mavrakis et al., 2006]. The second significant structural difference is a large loop from residue Leu117 to Glu127 in the VSV N protein, corresponding to the loop from residue Trp120 to residue Asp131 in the RABV N protein. This loop is below the N-terminal lobe of the VSV N protein whereas the same loop in the RABV N protein is below the RNA binding cavity. The third place is an insertion of three amino acids in the loop (residues Ile153-Thr158) in the RABV N protein that connects the helices 4 and 5 compared to the same loop (residues Thr147-Met149) in the VSV N protein. This loop is located at the mouth of the RNA binding cavity, and the insertion in the RABV N protein makes the RNA appeared to be more covered than that in the VSV N protein. Finally, there is an α–helix (residues Asp362-Tyr375) in the RABV N protein, which is absent in the VSV N protein. This helix is also present in the N protein of Borna disease virus [Rudolph et al., 2003], and that of the influenza A virus [Ye et al., 2006]. This helix precedes the P protein binding region in the RABV N protein [Dietzschold et al., 1987]. In addition, there is a short α–helix (residues Pro28-Ser35) in the VSV N protein not found in the RABV N protein.
Figure 1.
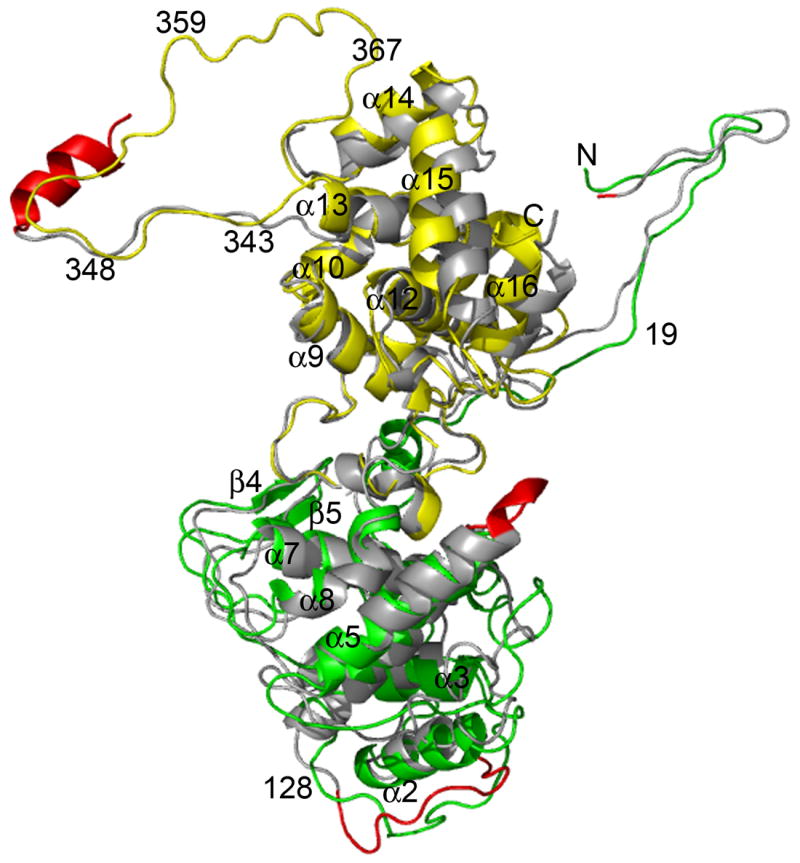
The superposition of structure of the VSV N protein with that of the RABV N protein shown as ribbon drawings. The N-terminal lobe of the VSV N protein is shown in green and the C-terminal lobe in yellow. The RABV N protein is shown in gray with regions that have structural differences in red. The view is into the RNA binding cavity. The N- and C-terminus, a number of residues, and a number of secondary elements are labeled according to the numbering of the VSV N protein. This protein cartoon and those following were prepared with the program PyMOL [Delano, 2002].
The RNA binding cavity is located between the conserved N-terminal and C-terminal domains (Figure 2). Bases in nucleotides 1–4 are stacked and face the opening of the cavity while bases in nucleotides 5, 7 and 8 are stacked and face the interior of the cavity. The base in nucleotide 6 is not involved in any stacking and is located at the tip of the cavity opening. In the VSV N protein, this base is sandwiched between the sidechains of Met149 and Arg408. In the RABV N protein, it is sandwiched between the larger loop (residues 153-Thr158) and the sidechain of Arg434. The larger loop in the RABV N protein shields the RNA much more than the sidechain of a methionine. The base in nucleotide 9 has a very different orientation in the VSV N protein from that in the RABV N protein, which could be the result of a changed curvature of the N-RNA complex (see below). The stacked based in nucleotides 1–4 are solvent accessible because they face the opening of the cavity, but the opening is too narrow to allow access to a large protein like the viral RNA polymerase. The stacked bases in nucleotides 5, 7 and 8 are accommodated in a large interior space that is hydrophobic. A number of the phosphate groups in the RNA are coordinated with sidechains of positively charged residues. Six such residues have been identified in the VSV N protein, including Arg143, Arg146 and Lys155 in the N-terminal domain, and Lys286, Arg317 and Arg408 in the C-terminal domain. However, the set of the positively charged residues that coordinate with the phosphate groups in the RNA is very different in the RABV N protein, contrary to the suggestion by Albertini et al., 2006 (Table 1). Only Arg143 and Arg408 in the VSV N protein are conserved in the RABV N protein, corresponding to Arg149 and Arg434, respectively. Lys155 in the VSV N protein is not aligned with Arg149 in the RABV N protein, but their sidechains do coordinate with the same phosphate group. The sidechain of Arg149 in the RABV N protein coordinates with two consecutive phosphate groups. The sidechain of Arg317 in the VSV N protein coordinates with the same phosphate group as that of Arg323 in the RABV N protein, but the two residues are not structurally aligned and their sidechains coordinate with the phosphate group from a different angle. Arg146 and Lys286 in the VSV N protein align with Lys152 and Lsy297 in the RABV N protein, respectively. However, the sidechains of the two residues in the RABV N protein do not coordinate with any phosphate group. In addition, Arg168 and Arg225 in the RABV N protein were found to coordinate with phosphate groups, but they align with Asp158 and Arg214 (not coordinating with any phosphate group) in the VSV N, respectively. It seems that in the RNA binding cavity, there are a large number of positively charged residues. In the N-RNA complex, only a subset of them coordinate with phosphate groups in the RNA, and a different set may be involved in different N proteins. In the VSV N protein, 6 out of 14 positively charged residues in the cavity participate in coordinating with the phosphate groups in the RNA. In the RABV N protein, 5 out of 9 residues participate. Another way to analyze the conserved features in the RNA binding cavity is to compare different strains of the N protein. There are two representative strains of VSV, Indiana and New Jersey serotypes. The VSV N protein from each strain has the same length of 422 amino acids and share a 68% identify in amino acid sequence. When the residues identical in both strains are mapped on the VSV N protein structure, these residues are concentrated in the RNA binding cavity (Figure 2c). All six positively charged residues that coordinate with the phosphate groups are conserved except that Lys155 is replaced by an arginine. This observation implies that the entire cavity contributes to the encapsidation of the RNA by not only providing positively charged residues but also a conserved shape of the cavity which is formed by the collection of all residues in the cavity.
Figure 2.
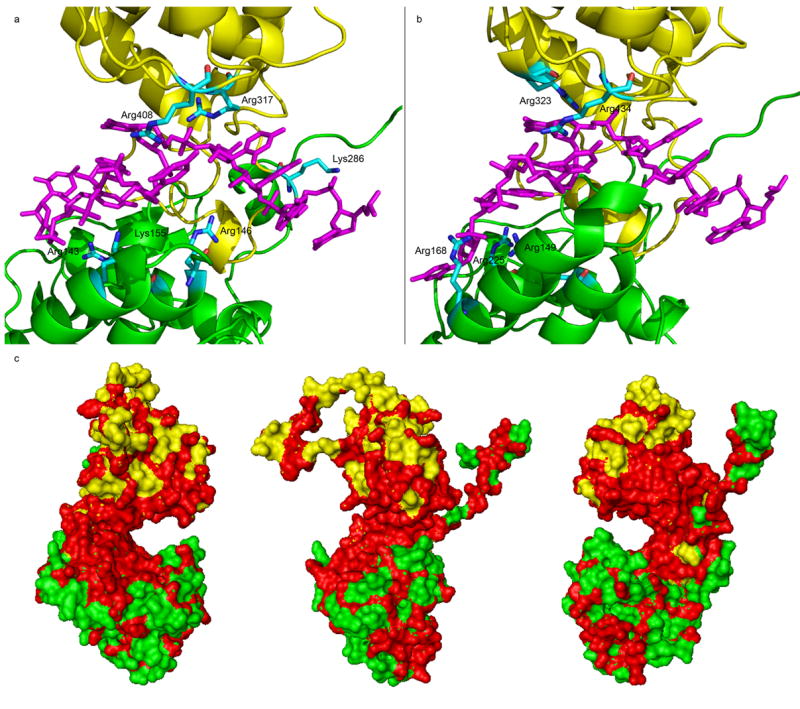
The RNA binding cavity. (a) The interactions of the positively residues with the RNA in the RNA binding cavity of the VSV N-RNA complex. The RNA is shown as a stick model (magenta) and the N-domain (green) and C-domain (yellow) of the N protein are shown as ribbons. The positively charged residues that coordinate with phosphate groups are labeled. (b) The interactions of the positively residues with the RNA in the RNA binding cavity of the RABV N-RNA complex. The scheme is the same as in (a). (c) The surface plot of identical residues (red) in the N protein of VSV Indiana and New Jersey serotypes. The panel in the center has the RNA binding cavity facing the reader. The other two views are from each side. The N-domain is colored green and the C-domain, yellow.
Table 1.
Residues coordinating with RNA
| Aligned residues in | |
|---|---|
| VSV N | RABV N |
| Arg143⇓ | Arg149 |
| Arg146 | Lys152|| |
| Lys155 | [Arg149]≠ |
| Asp158 | Arg168 |
| Arg214 | Arg225 |
| Lys286 | Lys297 |
| Arg312 | Arg323 |
| Arg317 | Thr328 |
| Arg408 | Arg434 |
Bold letters indicate that the sidechain of this residue coordinates with a phosphate group.
Italic letters indicate that the sidechain of the residues do not coordinate with any phosphate groups.
Residue Arg149 in the RABV N protein does not align with Lys155 in the VSV N protein even though their sidechains coordinates with the same phosphate group.
Extensive intermolecular interactions have been observed in the N-RNA complex spanning four neighboring N molecules. Three unique contacts cross N molecules are described in Green et al., 2006 as listed in Table 2, in addition to side-by-side contacts between the N molecules next to each other (Figure 3). Elimination of any one of the contacts will completely abolish the RNA encapsidation by the VSV N protein (Zhang et al., unpublished data). The three cross contacts are conserved in both N-RNA complexes, but the residues participating in these contacts are different. Each contact has a similar number of interactions in either the VSV N protein or the RABV N protein, with the contact II in the RABV N protein has slightly less interactions. The contacts in the RABV N protein appear to involve relatively more hydrophobic interactions, particularly sidechains of phenylalanines. In the side-by-side interactions of the C-terminal domain, the key contacts in the VSV N protein are that the sidechain of Tyr324 interacts with a hydrophobic pocket in the next N molecule, and other contacts around residue Tyr324. The salt bridge between the sidechain of Arg309 and that of Glu419 also contributes to this side-by-side interaction. In the RABV N protein, the tyrosine is replaced by a histidine (His336) whose sidechain may have hydrophobic interactions with a surface pocket in the next N molecule, as well as form a hydrogen bond with the carbonyl oxygen of Met319. The hydrogen bond between the carbonyl oxygen of Thr445 and the sidechain of Arg423, and the hydrophobic interactions by the sidechain of Phe350 with the surface pocket in the next N molecule could also contribute to the side-by-side interaction by the C-terminal domain in the RABV N protein. Both N proteins are capable of maintaining these contacts by engaging different amino acids, but in the same locations.
Table 2.
Residues related to intermolecular contacts
| VSV N | RABV N | ||||
|---|---|---|---|---|---|
| Interaction | Residue 1 | Residue 2 | Interaction | Residue 1 | Residue 2 |
| Contact I | Contact I | ||||
| H bond | O= of Ser2 | sidechain of Glu243 | Hydrophobic | sidechain of Phe8 | surface pocket |
| Salt bridge | sidechain of Arg7 | sidechain of Asp256 | H bond | sidechain of Asn12 | sidechain of Arg270 |
| H bond | sidechain of Lys17 | O= of Met262 | H bond | O= of Glu20 | sidechain of Arg254 |
| Hydrophobic | sidechain of Glu20 | sidechain of Phe273 | |||
| Contact II | Contact II | ||||
| H bond | sidechain of Ser340 | sidechain of Lys388 | H bond | -NH- of Gly353 | O= of Gly414 |
| H bond | O= of Ala342 | sidechain of Arg387 | H bond | -NH- of Thr354 | sidechain of Glu337 |
| H bond | -NH- of Asp343 | sidechain of Ser326 | Hydrophobic | sidechain of Phe355 | surface pocket |
| Hydrophobic | sidechain of leu344 | surface pocket | Hydrophobic | sidechain of Arg357 | surface pocket |
| H bond | -NH- of Ala345 | O= of Leu250 | Hydrophobic | sidechain of Phe359 | surface pocket |
| Hydrophobic | sidechain of Phe348 | surface pocket | |||
| Hydrophobic | sidechain of Lys354 | sidechain of Leu379 | |||
| Contact III | Contact III | ||||
| H bond | -NH- of Lys6 | O= of Cys349 | H bond | -NH- of Lys9 | O= of Phe360 |
| H bond | O= of Lys6 | -NH- of Cys349 | H bond | O= of Lys9 | -NH- of Phe360 |
| H bond | -NH- of Ile8 | O= of Gln347 | Salt bridge | sidechain of Lys9 | sidechain of Glu363 |
| Hydrophobic | sidechain of Val10 | sidechain of Phe359 | |||
Figure 3.
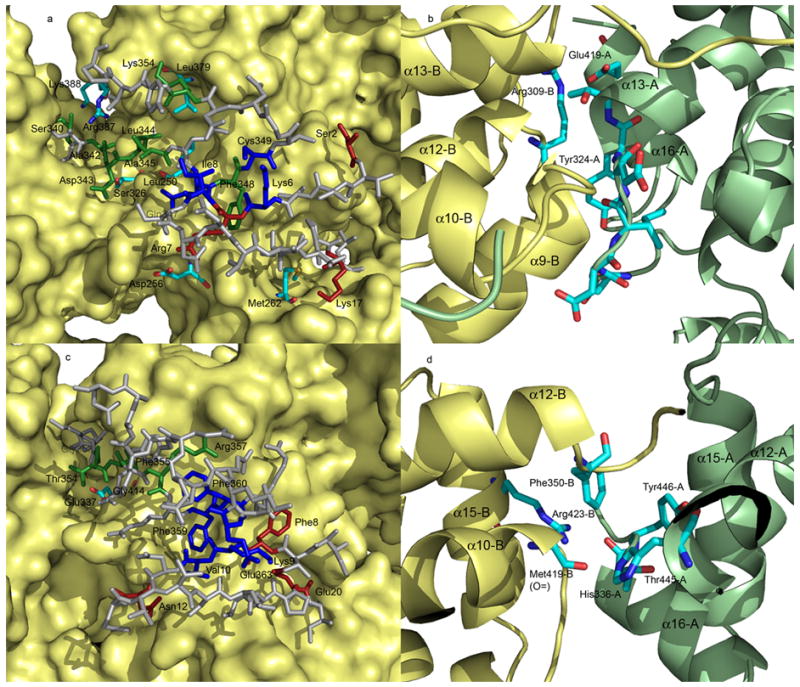
Intermolecular interactions. (a) The cross molecule contacts in the VSV N-RNA complex. The main body of the N molecules is presented as surface rendering (yellow). The residues in the N-terminal arm and the extended loop that are involved in contact I are in red, contact II, green, and contact III, blue. Residues that count-interact the above residues except for those forming surface pockets are colored by elements. Residues are labeled with –A indicating from molecule A, and –C indicating from molecule C. Other label residues are from molecule B. The remaining residues in the N-terminal arm and the extended loop are in gray. (b) The side-by-side interactions of the C-terminal domains. Molecule A is in green and molecule B in yellow. Residues involved in the interactions are colored by element and labeled accordingly. A number of secondary elements are also labeled. (c) The cross molecule contacts in the RABV N-RNA complex, and (d) the side-by-side interactions of the C-terminal domains. The scheme is the same as in (a) and (b), respectively.
The RNP is condensed into a superhelix with 32 to 38 N molecules per turn inside the virion (Z.H. Zhou, personal communication), and becomes linear after uncoating in the cytoplasm. The current N-RNA complexes for which the crystal structures were solved contain 10 and 11 N molecules, respectively. Comparisons of the two ring complexes may explain how the RNP may assume different conformations at a different curvature between the N molecules. The angle between two neighboring molecules in the VSV N-RNA ring is 144°, and it is 147.3° in the RABV N-RNA ring. In the virion, this angle becomes between 168.8° and 170.5°. In the linear form of the RNP, this angle does not seem to be 180° because the electron microscopy image of the RNP showed that it has coil features [Barge et al., 1993]. The change from the VSV N-RNA ring to the RABV N-RNA ring represents an increase in the curvature. There are two major structural changes associated with the increase of the curvature. The first change is the base stacking of the RNA. In the VSV N-RNA ring, the base in nucleotide 9, located at the interface of two neighboring N molecules, does not stack with any other bases (Figure 4a). Instead, the sidechain of Tyr215 stacks with the base in nucleotide 1. When the curvature increases as observed in the RABV N-RNA ring, nucleotide 9 changes its conformation so that the base in nucleotide 9 is now stacked with that in nucleotide 1. Since the distance for encapsidating 9 nucleotides per N molecule is fixed in the N-RNA complex, addition of an additional stacking base causes the RNA to be more compacted. Nucleotide 9 appears to be the flexible point in the RNA to accommodate the change in the curvature by adjusting the conformation of nucleotide 9. Nucleotide 1–8 is likely to move with the N molecule as a rigid body in any conformational change of the RNP. Another major change is the last three helices near the C-terminus. When the space between the two neighboring N molecules increases upon an increase of curvature, the three helices that follow the P protein binding loop move toward the gap between the two N molecules as observed in the RABV N-RNA ring compared to that in the VSV N-RNA ring (Figure 4b). This movement covers the top of the encapsidated RNA and keeps the protein cage surrounding the RNA continuous.
Figure 4.
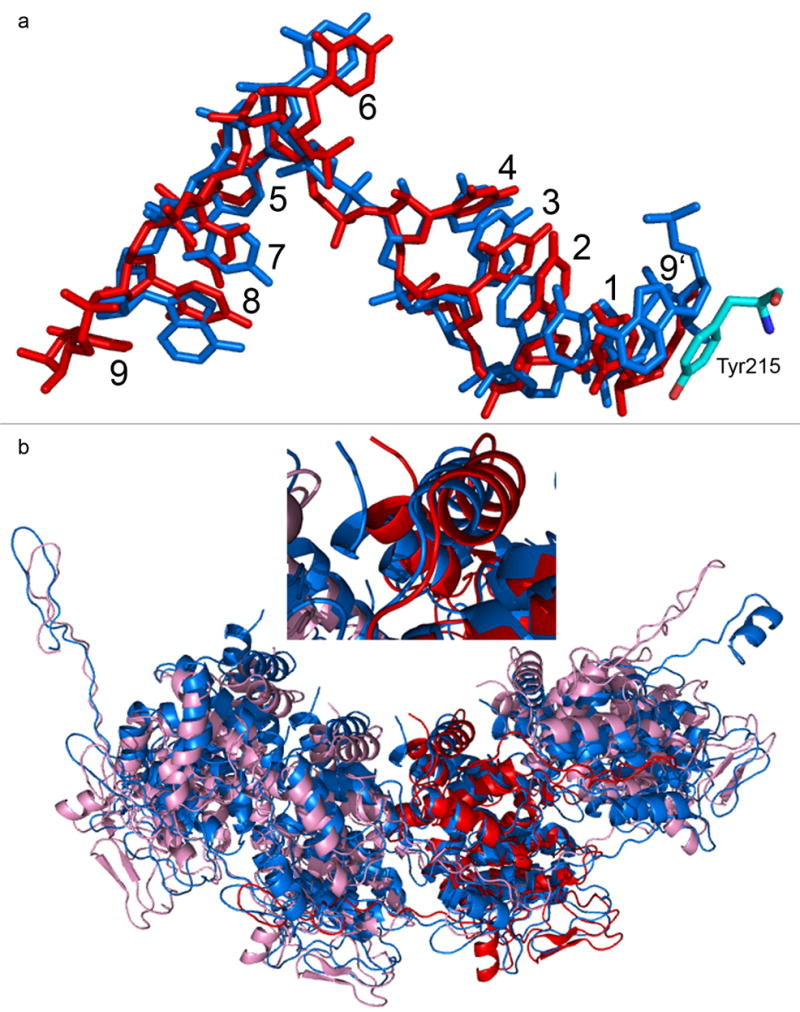
Structural changes related to the RNP curvature. (a) the conformation of the nine nucleotides per N molecule in the VSV N-RNA ring (red), that in the RABV N-RNA ring (blue), and their superposition. The sidechain of Tyr215 that stacks with the RNA in the VSV N-RNA ring was shown as colored by element. Nucleotides are numbered according to [Green et al., 2006]. Number 9′ indicates the nucleotide belongs to the previous protomer. (b) The superposition of the VSV N-RNA ring (pink) with that of the RABV N-RNA ring (blue). Only four unique N molecules were selected from each ring. One N molecule (red in the VSV N-RNA ring) was selected as the reference of the superposition with the program DALI [Holm & Park, 2000]. The inset is the close-up view of the three helices near the C-terminus showing the movement of the C-terminus (blue) in the RABVN-RNA complex toward the interface between two N molecules.
The comparisons of the N protein from different rhabdoviruses showed that the protein fold is highly conserved despite their low homology in amino acid sequence. A limited number of local structural changes are found outside of the primary fold of the two structural domains. The RNA binding cavity is the highest conserved feature in the N protein as demonstrated by the two representative strains of VSV. However, the N protein from two different members of rhabdovirus family, VSV and RABV, may use different amino acids to encapsidate the RNA that has the same conformation in the cavity of both N proteins. This emphasizes that it is the shape of the cavity that packages the RNA in the N-RNA complex. The interactions to bind the RNA may be rendered by different residues lining the cavity, including both positively charged and uncharged residues. Positively charged residues are not highly conserved in the structure as previously expected. The cross molecule interactions that are required for RNA encapsidation are conserved by the different N proteins, but also through a different set of amino acids. The N protein and the RNA have sufficient flexibilities to adopt different RNP conformations that are present at different steps in the virus replication cycle. The major point of flexibility is around nucleotide 9 that is located at the interface of two neighboring N molecules. These general characteristics observed in the N-RNA complex of rhabdoviruses are likely to be common in the RNP of other negative strand RNA viruses, but the amino acids involved in the interactions may be very different.
Acknowledgments
We thank Dr. Z. Hong Zhou and Mr. Peng Ge for stimulating discussions. The work is supported in part by a grant from NIH AI050066.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Barge A, Gaudin Y, Coulon P, Ruigrok RW. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J Virol. 1993 Dec;67(12):7246–53. doi: 10.1128/jvi.67.12.7246-7253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. The PyMOL User’s Manual. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- Dietzschold B, Lafon M, Wang H, Otvos L, Jr, Celis E, Wunner WH, Koprowski H. Localization and immunological characterization of antigenic domains of the rabies virus internal N and NS proteins. Virus Res. 1987 Aug;8(2):103–25. doi: 10.1016/0168-1702(87)90023-2. [DOI] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–60. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Mehouas S, Real E, Iseni F, Blondel D, Tordo N, Ruigrok RW. Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology. 2006 Jun 5;349(2):422–9. doi: 10.1016/j.virol.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Kraus I, Dickmanns A, Eickmann M, Garten W, Ficner R. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11:1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Wang Y, Cowley JA, McWilliam SM, Prehaud CJ. Structural and antigenic analysis of the nucleoprotein of bovine ephemeral fever rhabdovirus. J Gen Virol. 1994 Aug;75(Pt 8):1889–99. doi: 10.1099/0022-1317-75-8-1889. [DOI] [PubMed] [Google Scholar]
- Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]


