Abstract
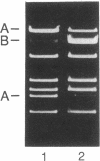
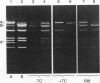
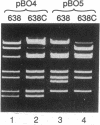
pBF4 is a 42-kb R plasmid from Bacteroides fragilis which transfers clindamycin resistance (Clr) independently of the chromosomal tetracycline resistance (Tcr) transfer element. We have found that this plasmid exists in two nonequimolar conformations, A and B. These forms differ by an inversion of approximately 11.5 kb which does not involve the repeated DNA sequences previously mapped on the plasmid. The presence of chromosomal tetracycline resistance conjugal elements influences the relative amounts of the two conformations: induction with tetracycline shifts the dominant form from B to A.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I., Ambrosio L. The invertible segment of bacteriophage Mu DNA determines the adsorption properties of Mu particles. Nature. 1978 Feb 9;271(5645):575–577. doi: 10.1038/271575a0. [DOI] [PubMed] [Google Scholar]
- Futcher A. B. The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast. 1988 Mar;4(1):27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Bouic K., Matthews B. Genetic transfer systems in Bacteroides: cloning and mapping of the transferable tetracycline-resistance locus. Mol Microbiol. 1989 Nov;3(11):1617–1623. doi: 10.1111/j.1365-2958.1989.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Homology between clindamycin resistance plasmids in Bacteroides. Plasmid. 1984 May;11(3):268–271. doi: 10.1016/0147-619x(84)90035-0. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Stalker D., Davis C. E. Genetic analysis of clindamycin resistance in Bacteroides species. J Infect Dis. 1983 Mar;147(3):551–558. doi: 10.1093/infdis/147.3.551. [DOI] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Komano T., Funayama N., Kim S. R., Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990 May;172(5):2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T., Kubo A., Kayanuma T., Furuichi T., Nisioka T. Highly mobile DNA segment of IncI alpha plasmid R64: a clustered inversion region. J Bacteriol. 1986 Jan;165(1):94–100. doi: 10.1128/jb.165.1.94-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kusukawa A., Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988 Jul;213(1):30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- Magot M., Fayolle F., Privitera G., Sebald M. Transposon-like structures in the Bacteroides fragilis MLS plasmid plP 410. Mol Gen Genet. 1981;181(4):559–561. doi: 10.1007/BF00428754. [DOI] [PubMed] [Google Scholar]
- Matthews B. G., Guiney D. G. Characterization and mapping of regions encoding clindamycin resistance, tetracycline resistance, and a replication function on the Bacteroides R plasmid pCP1. J Bacteriol. 1986 Aug;167(2):517–521. doi: 10.1128/jb.167.2.517-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- Odelson D. A., Rasmussen J. L., Smith C. J., Macrina F. L. Extrachromosomal systems and gene transmission in anaerobic bacteria. Plasmid. 1987 Mar;17(2):87–109. doi: 10.1016/0147-619x(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Privitera G., Sebald M., Fayolle F. Common regulatory mechanism of expression and conjugative ability of a tetracycline resistance plasmid in Bacteroides fragilis. Nature. 1979 Apr 12;278(5705):657–659. doi: 10.1038/278657a0. [DOI] [PubMed] [Google Scholar]
- Salyers A. A., Shoemaker N. B., Guthrie E. P. Recent advances in Bacteroides genetics. Crit Rev Microbiol. 1987;14(1):49–71. doi: 10.3109/10408418709104435. [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Getty C., Gardner J. F., Salyers A. A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986 Mar;165(3):929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Salyers A. A. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J Bacteriol. 1988 Apr;170(4):1651–1657. doi: 10.1128/jb.170.4.1651-1657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J Bacteriol. 1985 Mar;161(3):1069–1073. doi: 10.1128/jb.161.3.1069-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Spiegel H. Transposition of Tn4551 in Bacteroides fragilis: identification and properties of a new transposon from Bacteroides spp. J Bacteriol. 1987 Aug;169(8):3450–3457. doi: 10.1128/jb.169.8.3450-3457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Welch R. A., Macrina F. L. Two independent conjugal transfer systems operating in Bacteroides fragilis V479-1. J Bacteriol. 1982 Jul;151(1):281–287. doi: 10.1128/jb.151.1.281-287.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Shimell M. J., Malamy M. H. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis. J Bacteriol. 1982 Aug;151(2):686–691. doi: 10.1128/jb.151.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J., Silverman M., Hilmen M., Simon M. Recombinational switch for gene expression. Science. 1977 Apr 8;196(4286):170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]