Abstract
Background
The development of post-genomic methods has dramatically increased the amount of qualitative and quantitative data available to understand how ecological complexity is shaped. Yet, new statistical tools are needed to use these data efficiently. In support of sequence analysis, diversity indices were developed to take into account both the relative frequencies of alleles and their genetic divergence. Furthermore, a method for describing inter-population nucleotide diversity has recently been proposed and named the double principal coordinate analysis (DPCoA), but this procedure can only be used with one locus. In order to tackle the problem of measuring and describing nucleotide diversity with more than one locus, we developed three versions of multiple DPCoA by using three ordination methods: multiple co-inertia analysis, STATIS, and multiple factorial analysis.
Results
This combination of methods allows i) testing and describing differences in patterns of inter-population diversity among loci, and ii) defining the best compromise among loci. These methods are illustrated by the analysis of both simulated data sets, which include ten loci evolving under a stepping stone model and a locus evolving under an alternative population structure, and a real data set focusing on the genetic structure of two nitrogen fixing bacteria, which is influenced by geographical isolation and host specialization. All programs needed to perform multiple DPCoA are freely available.
Conclusion
Multiple DPCoA allows the evaluation of the impact of various loci in the measurement and description of diversity. This method is general enough to handle a large variety of data sets. It complements existing methods such as the analysis of molecular variance or other analyses based on linkage disequilibrium measures, and is very useful to study the impact of various loci on the measurement of diversity.
Background
The exponential increase in sequencing abilities is modifying the way genetic diversity is assessed. For instance, multilocus sequencing (MLS) now allows the estimation of genetic relatedness among microorganisms for both housekeeping genes and accessory genes such as virulence or symbiotic determinants [1]. Thus, several publications reported complex MLS schemes studying more than ten genes located in different genomic regions and involved in various metabolic pathways. These studies have indicated the influence of various parameters, such as recombination rate [2] or epidemiological traits [3], on the diversification of bacterial populations. Furthermore, recent progress in sequencing technologies suggests that still more and more sequence data will be available to study questions related to community ecology in the near future [4]. New statistical methodologies should therefore be developed to deal with the complexity of data sets that will be produced. One of the main problems raised by the increase in sequence information is the assessment of congruence among population structures depicted by different molecular markers [5]. In bacterial lineages, especially for those in which sex is common, the diversity of each locus could be shaped by the gain/loss of genes, gene flow boundaries and specific selective pressures [6]. The problems which can arise from the overall analysis of a MLS data set in which loci do not share congruent evolutionary constraints include, among others, misleading inferences of genetic relatedness and phylogenetic relationships [7] or overestimation of linkage disequilibrium [8].
Bacterial isolates which are characterized by MLS usually belong to several genetic groups (i.e. species or populations) which can be defined according to the sampling strategy or according to more refined methodologies [9]. For each locus of a MLS data set, the different sequence types recovered are called alleles. In this context, the properties of the data set can be summarized by two sets of matrices. The first set includes G matrices {F1,..., Fg,..., FG}, in which G is the number of loci. Each of these matrices contains the frequencies of the different alleles recovered at a given locus among the populations under study. The dimensions of these matrices are thus (ρ1, r), ..., (ρg, r), ..., (ρG, r), in which ρg is the number of alleles observed at locus g and r is the number of populations delineated. The second set also includes G matrices called {D1,..., Dg..., DG}, which contain the pairwise genetic distances between the alleles observed at locus g. Usually, the information contained within these two sets of matrices are analyzed independently using respective population genetic statistics (i.e. diversity indices and differentiation measures) and phylogenetic methods. Yet, while it is possible to perform analyses over all loci in either a population genetic or a phylogenetic framework, few methodologies are available to assess the congruence of the information obtained from different loci. In particular, a comparison of the patterns revealed by differentiation measures among the populations sampled, i.e. population structure, is a problematic issue.
Multivariate analysis is an interesting methodological way to approach this problem. For instance, Moazami-Goudarzi and Laloë [5] have proposed a two-step procedure to test the dissimilarity in population structures revealed by different microsatellite loci. Although this analysis can be used to test the similarity of population differentiations inferred from a set of markers, it can be noted that: i) it can not be used to describe population structures, and ii) genetic divergence among alleles are not taken into account, while these can be quite informative. Consequently, further improvements should be considered since alternative statistical approaches are available [10]. In this context, the aim of this survey is to propose a new procedure called multiple double principal coordinate analyses (mDPCoA). The mDPCoA aims at comparing inter-population structures provided by the different markers of a MLS scheme. Firstly, a pattern of population differences is obtained for each MLS marker using a double principal coordinate analysis (DPCoA) which is a recently developed ordination method which takes into account both the frequency of alleles and their genetic divergence [11] (see Eckburg et al. [12] and Bik et al. [13] for applications of this method to the analysis of bacterial diversity). Secondly, population patterns are compared using three different methods: the Multiple Co-inertia Analysis [14], STATIS [15], and the Multiple Factorial Analysis [16]. Finally, a permutation procedure can be used to test the pairwise correlation among MLS markers. These analysis pipelines have been used on either simulated or published MLS data sets to check the accuracy and the relevance of the procedures. The results obtained illustrate the ability of this methodology to make inferences on various features of populations under study.
Results
Algorithms of multiple Double Principal Coordinate Analysis
Computations were performed using new functions and functions implemented in the ade4 [17] and ape [18] packages written in the R software [19] [see Additional file 1]. A manual describing the use of the different functions is supplied [see Additional file 2].
Let {F1,..., Fg,..., FG be the set of matrices of type alleles × populations, containing the frequencies of alleles in the populations for the G loci, {D1,..., Dg,..., DG} be the set of matrices containing the distances among alleles, Br be the diagonal matrix containing the population weights (the weight of a population is the proportion of individuals drawn from this population), and be the diagonal matrix containing the allele weights for the gth locus (the weight of an allele is its frequency over all the populations studied). The matrices of distances must be Euclidean [20], which is obtained with, for example, either Lingoes [21] or Cailliez [22] correction.
For a single locus g, the analysis of the among-population diversity corresponds to a DPCoA, which results in three main steps:
1. Defining a Euclidean space composed by principal axes of the distances among the alleles. The coordinates of the alleles in this space are in Rg such that: , where is a projector which proceeds to weighted centering, with the ρg × ρg matrix of identity and a ρg × 1 vector of units. That is to say, is the matrix centered by rows and columns;
2. Positioning, in this space, the populations at the centroid of the alleles they possess. The coordinates of the populations, in this space, are in Cg such that: ;
3. Proceeding to the singular value decomposition of the triplet (Cg, , Br), where μg is the number of principal axes for the alleles of the gth locus. This third step leads to a set of positive eigenvalues, in a diagonal (νg × νg) matrix Ψg, and to a base of orthonormal eigenvectors, in a (r × νg) matrix Vg, defining the new Euclidean space. The eigenvectors constitute the principal axes of the distances among populations. In this new space, which is the DPCoA space, the coordinates of the alleles are in Xg = RgVg, and the coordinates of the populations in Yg = CgVg.
A consideration of the set of all the loci leads thus to G triplets
Our objective being to evaluate the consistency among the patterns of inter-population diversity provided by each locus, considering evolutionary distances among alleles, we had to find a Euclidean space allowing the direct comparison among the individual DPCoA analyses. We evaluated three alternative solutions taken from the K-table multivariate analysis: the multiple co-inertia analysis (MCoA) [14], STATIS [15] and the multiple factorial analysis (MFA) [16].
DPCoA and Multiple Co-inertia analysis
The Multiple Co-inertia Analysis applied to the triplets .
can be viewed as follows:
The main step is the definition of a set of axes , for 1 ≤ k <K, and 1 ≤ g ≤ G, normalized in each space , which will serve to position the populations according to each individual locus, and K unique variables v[k], for 1 ≤ k <K, Dr-normalized in ℝr, which may be used to synthesize the information provided by the G loci. This definition is done by maximizing
, given that
and for all k, l (1 ≤ k <l), and all g (1 ≤ g ≤ G).
The value πg is a weight attributed to the triplet (Yg, , Br) so as to homogenize the impact of each triplet in the multiple analysis. We use πg equal to the inverse of the inertia of the triplet (Yg, , Br), sum of all its eigenvalues. Let Ug be the matrix and V the matrix [v[1]|...|v[k]|...|v[k]]. The individual analyses can be projected on the MCoA space. In this space, it is possible to compare the coordinates of the populations according to the consensus of the information provided by the different loci to the coordinates of the populations obtained from each locus. While V contains the consensual coordinates of the populations, the coordinates at which the gth locus positions the populations are obtained from . Because , the matrix positions the alleles of the gth locus, so that each population is at the centroid of its allelic composition. However, to compare the individual analyses with the compromise, it is better to Dr-normalize and because V is by definition Dr-normalized.
DPCoA and STATIS
The STATIS analysis applied to implies the calculation of a degree of correlation among the triplets, the so-called Rν coefficient. The matrix
is at the core of our application of STATIS because it is symmetrical and its dimensions are similar for all the triplets, whereas the dimensions of Yg change. The definition of Rν is
where
The pairwise calculation of Rν leads to a square matrix describing the correlations among the loci. With its eigenvalue decomposition, it is possible to describe the correlation pattern, called the interstructure. Its first eigenvector α = (α1,..., αg,..., αG) is positive and maximizes the quantity where . STATIS uses these properties to define a matrix
whose eigenanalysis, E = UΛUt, leads to the best compromise of the population pattern over the G loci. Note that . According to this compromise, the coordinates of the populations are in . Owing to Lavit et al. [15], the G individual population patterns corresponding to the locus considered independently can be obtained. The coordinates of the ith populations according to the gth locus are the elements of the ith row of . Given that , the rows of the matrix position the alleles of the gth locus, so that each population is at the centroid of its allelic composition.
DPCoA and Multiple Factorial Analysis
The MFA is the Principal Component Analysis (PCA) of the global matrix
| YTOT = [π1Y1|...|πgYg|...|πGYG]: |
The global coordinates of the populations synthesizing the information given by all the loci are in YTOTU. The coordinates at which the gth locus positions the populations are in
Because , the matrix positions the alleles of the gth locus, so that each population is at the centroid of its allelic composition.
Relationships between the multiple DPCoA and the measurement of diversity
Consider for the two next paragraphs, only one locus – the locus g. The DPCoA is centered around a diversity index called "nucleotide diversity" by Nei and Li [23], or "quadratic entropy" by Rao [24], and which is at the core of the Analysis of Molecular Variance (AMOVA) [25-27]:
In this formula, g designates the gth locus, ρg is the number of different alleles observed for that locus, is the vector containing the relative frequencies of the alleles in the ith population, so that pki is the frequency of the allele k in the ith population, and is the distance among the alleles k and l of the gth locus. The DPCoA uses a decomposition of this diversity component defined by Rao [27]:
| HTOTAL, g({μi},{pi}) = HINTRA, g({μi},{pi}) + HINTRA, g({μi},{pi}), |
where
and
where .
In the first step of the DPCoA, all the points (i.e. alleles and populations) are in a space called "common space" [11]. In this common space, the inertia (i.e. variance) of the allele points weighted by pi is equal to Hg(pi), the diversity of the population i, according to locus g. The inertia of all the allele points weighted by is equal to HTOTAL, g, the total diversity of the data set. Finally, the inertia of all the population points weighted by μ = (μ1,..., μi,..., μr) is equal to HINTER, g, the component of diversity among populations [11]. At the end of the DPCoA analysis, all the points are projected in a subspace which optimizes the representation of the differences among populations. In this subspace, only HINTER, g is maintained, which is thus the focus of the analysis: optimally displaying the diversity among populations.
Consequently, the multiple DPCoA allows us to optimize the description of diversity among populations obtained with several loci. The first goal of this method is to describe the differences in population patterns across the loci, hence studying the congruence among loci. Another objective may be to erase these differences and provide a compromise population pattern revealed by the majority of the loci. The DPCoA-STATIS is advocated for this purpose. Concerning the measurement of diversity, when several loci are considered to measure diversity, the sum or average of the diversity components over the loci is currently used as a global measure of diversity [see for example [28,29]]. With such processes, the weights given to the loci for the sum or averaging are uniform. We have just shown that STATIS provides optimal locus weights for the calculation of the component of diversity among populations. The great advantage of these multivariate analyses is that visualization of the differences among loci is possible so that one can assess the relevance of using average information over loci, whether these means are weighted or not.
Associated tests
We performed both Mantel and Rν tests to evaluate the significance of the differences in population patterns among loci. For each locus, distances among populations are calculated with the inter-population diversity HINTER, g({μi}:{pi}) according to Nei and Li [23] and Rao [24,27]. We just said that this statistic is at the core of the DPCoA. As we apply formula (HINTER, g) in a pairwise fashion, the distance between population i and population j for locus g is μiμjdpop, g(pi, pj). We choose μiμjdpop, g(pi, pj) and not simply dpop, g(pi, pj) to take into account differential sample sizes, exactly in the way that we considered them in ordination procedures. The Mantel test calculates correlations among the raw distance measures, while the Rν test compares principal coordinates obtained by PCoA. Rν correlations are always higher than Mantel correlations because their values lie between 0 and 1, while Mantel correlation values lie between -1 and 1.
Application to simulated and real data sets
We used the following procedure to test the methodologies presented above based on simulated and real data sets. First, pairwise correlations among loci by Mantel and/or Rν tests were assessed to define groups of consistent loci. At this step, atypical loci can be identified. Then mDPCoA was performed to describe both the compromise population structure and the differences among groups of loci. Finally, we describe the connections between the observed structures and ecological, evolutionary or functional data.
Application to a simulated data set
Simulation process
In order to assess the efficiency of the present method, simulated sequence data sets, which illustrate various population structures, were obtained assuming linkage equilibrium among loci. Assuming recombination, the different markers can indeed have different histories and thus different population structures. Moreover, if every marker has an independent history, finding similarities and differences among their genetic structures would be more difficult. Using SIMCOAL 2.0 [30] we considered a one-dimensional stepping stone model with eight populations of constant size [31]. The eight populations evolved 106 generations after emerging from a single ancestral population. For each population, 60 individuals were sampled out of 10000 individuals. In this context, we simulated DNA sequence evolution of ten loci of 300 base pairs under a Jukes and Cantor model [32] assuming a mutation rate of 5 × 10-6. The stepping stone model allows migration between adjacent populations: for example, at time t, the population 4 can exchange individuals with populations 3 or 5, but not with other populations. We chose the following migration rates: 5 × 10-2, 10-2, 5 × 10-3, 10-3, 5 × 10-4, 10-4, 5 × 10-5, 10-5, 5 × 10-6. We also simulated an eleventh locus that reveals a different population structure. For this locus, we assumed no migration between odd populations (i.e. populations 1, 3, 5, 7) and even populations (i.e. populations 2, 4, 6, 8) and a migration rate of 10-3 among odd or even populations, with other parameters kept unchanged. Such a simulation resulted in two clades of alleles which are obviously divergent, the first clade being specific to some populations (e.g. odd ones), the second clade being specific to other populations (e.g. even ones). Such genetic structure can be observed in case of either balancing/disruptive selection [e.g. [33]] or horizontal transfer of an outlier allele [e.g. [7]].
We applied the mDPCoA approach first on the complete data set, second on the allele distances only and then taking into account just the allele frequencies. We evaluated the intensity of inter-population structure by measuring the AMOVA ϕST parameter [25].
Results
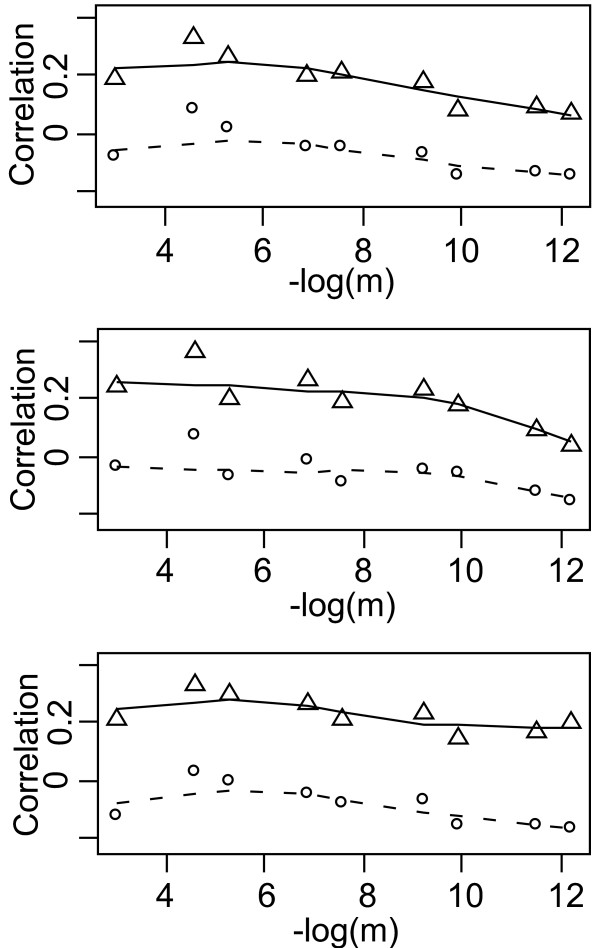
The correlations among locus 11 and the ten other loci are very low and not significant as expected (Figure 1). Thus, we correctly identified the atypical locus. These correlations decrease when migration rate decreases. Test statistics based on both the Mantel correlation and the Rν correlation between the atypical locus and other loci clearly behave in a similar way, and results are hardly changed when removing allele frequencies or distances.
Figure 1.
Mantel and Rv correlations between atypical and other loci in the simulated data set. The parameter m is the migration rate of the simulated linear stepping stone. Each statistic is calculated and averaged between the atypical locus and the first 10 loci submitted to a stepping stone model, A) with both allele frequency and distance information, B) with allele distances without allele frequencies, C) with allele frequencies without allele distances. Plain lines with triangle-shaped symbols mark the average Rν correlation values, while the broken lines with open circles indicate the average Mantel correlation values.
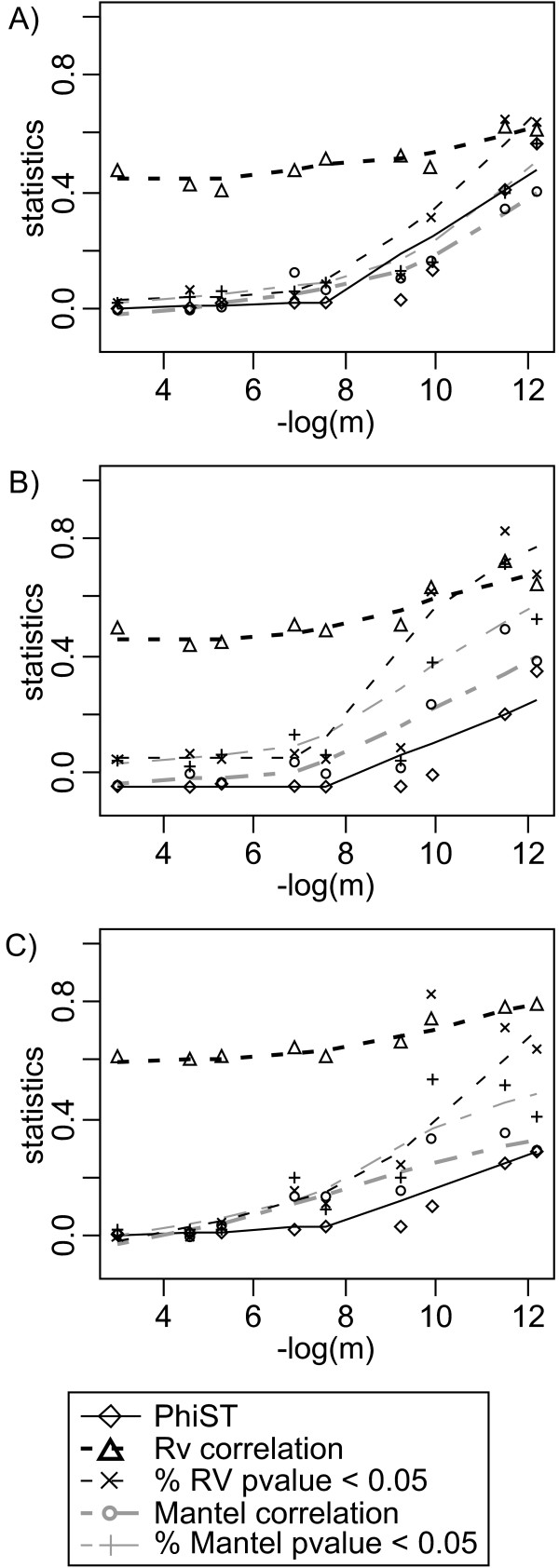
Regarding the correlation tests among the 10 loci submitted to the stepping stone model, the inter-population structure measured by the AMOVA ϕST parameter increases slightly when the migration rate decreases from 5 × 10-2 to 5 × 10-4 and then increases very quickly (Figure 2). Values of the Mantel correlation, the percent of significant tests according to the Mantel correlation and the percent of significant tests according to the Rν correlation are three parameters correlated with ϕST, especially when using both allele frequency and allele divergences. The raw value of the Rν correlation is steadier. These results show that a non-significant correlation may be due to either an absence of genetic structure (e.g. no differentiation among populations) or reliable differences in the inter-population structures revealed by the different loci. The graphical analysis completed by ϕST values will help to reach a conclusion between the two alternatives.
Figure 2.
Mantel and Rv correlations among the ten first loci in the simulated data set. The parameter m is the migration rate of the simulated linear stepping stone. Each statistic is calculated on 10 loci submitted to this stepping stone model, A) with allele frequency and distance information, B) with allele distances without allele frequencies, C) with allele frequencies without allele distances. Symbol legends are given at the bottom of the graphs.
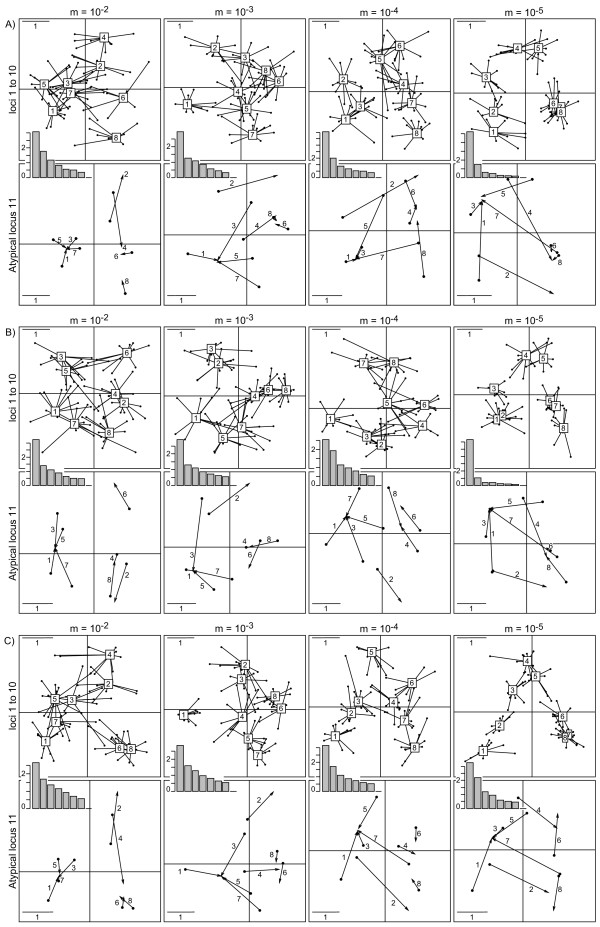
Regarding the mDPCoA, we present below the results of the DPCoA-MCoA approach, which we expected to provide a description of the difference among the ten first loci and the eleventh, atypical locus (Figure 3; to limit the size of the Figure 3, only the results for migration rates 10-2, 10-3, 10-4 and 10-5 are shown since intermediate migration rates revealed intermediate inter-population structure). Indeed, for migration rates higher than 10-2, where no inter-population structure was highlighted in the previous paragraph, the atypical locus takes the first axis of the compromise analysis, which therefore distinguishes odd from even populations. With a migration rate of 10-3, the stepping stone model interacts with the structure provided by locus 11; the 10 first loci with a stepping stone model take the first axis and locus 11 roughly takes the second axis. With a migration rate lower than 10-3, the first two axes of the DPCoA-MCoA only represent the stepping stone model. Whatever the migration rate, the projection of the individual loci on the DPCoA-MCoA factorial axes emphasizes locus 11's special status (Figure 3). This last result is also emphasized by specific results of the DPCoA-STATIS approach as interstructures. With a migration rate equal to 5 × 10-4 or lower, the structure is very clear with either complete or incomplete data on allele composition.
Figure 3.
Application of the DPCoA-MCoA to the simulateddata set. The parameter m is the migration rate of the simulated linear stepping stone. The DPCoA-MCoA was applied on the simulated data set, A) with allele frequency and distance information, B) with allele distances without allele frequencies, C) with allele frequencies without allele distances. Each figure A) B) and C) comprises two series of four subfigures. In the first row, for each locus the compromise pattern of differences among populations (Numbers in boxes) is given with lines relating the compromise to the ten first loci submitted to the stepping stone model. In the second row, for each locus the compromise pattern of population differences is also given at the beginning of the arrows, and this time, the arrows point at the position of each population according to the atypical locus. The longer the arrow, the more different the pattern inferred by the atypical locus from the compromise pattern. Eigenvalue barplots are provided for analyses A), B), and C).
Application to the description of Sinorhizobium species diversity
The data set
In order to test the efficiency of the procedures we proposed, we needed a real data set which should give simple and explicit results but which could also encompass the features of complex MLS data sets. We chose to focus on nitrogen fixing bacteria belonging to the genus Sinorhizobium (Rhizobiaceae) associated with the plant genus Medicago (Fabaceae). The data set we chose is a combination of two data sets fully available online from GenBank and published in two recent papers [8,34]. The complete sampling procedure is described in the two papers and summarized in an additional file [see Additional file 3]. Based on the sampling scheme, we delineated six populations according to geographical origin (France: F, Tunisia Hadjeb: TH, Tunisia Enfidha: TE), the host plant (M. truncatula or similar symbiotic specificity: T, M. laciniata: L), and the taxonomical status of bacteria (S. meliloti: mlt, S. medicae: mdc). Each population will be called hereafter according to the three above criteria, e.g. THLmlt is the population sampled in Tunisia at Hadjeb from M. laciniata nodules which include S. meliloti isolates. S. medicae interacts with M. truncatula while S. meliloti interacts with both M. laciniata (S. meliloti bv. medicaginis) and M. truncatula (S. meliloti bv. meliloti) [35,36]. The numbers of individuals are respectively 46 for FTmdc, 43 for FTmlt, 20 for TETmdc, 24 for TETmlt, 20 for TELmlt, 42 for THTmlt and 20 for THLmlt [see Additional files 4, 5, 6, 7].
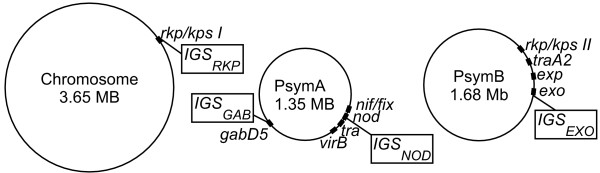
Four different intergenic spacers (IGS), IGSNOD, IGSEXO, IGSGAB, and IGSRKP, distributed on the different replication units of the model strain 1021 of S. meliloti bv. meliloti (Figure 4) had been sequenced to characterize each bacterial isolate (DNA extraction and sequencing procedures are described in an additional file [see Additional file 3]). It is noteworthy that the IGSNOD marker is located within the nod gene cluster and that specific alleles at these loci determine the ability of S. meliloti strains to interact with either M. laciniata or M. truncatula [37].
Figure 4.
Location of genetic markers on the genome of Sinorhizobium meliloti strain 1021. Gene clusters located nearby each genetic marker are indicated by black boxes. It is noteworthy that the IGSNOD marker is located near genes involved in symbiotic specificity (nod genes), symbiotic efficiency (nif/fix genes), secretion (virB gene) and conjugation (tra genes). IGSRKP and IGSEXO are located near genes involved in the synthesis of surface polysaccharides, which are also involved in the symbiotic interaction. IGSGAB is physically close to genes involved in secondary metabolic pathways.
For each locus, we selected a model of evolution using the software PHYML [38] and its R interface provided by ape [18,19]. This software compares the models by likelihood ratio tests. When several models were not significantly different according to a χ2 test we selected the model with the smallest number of parameters. From this procedure, we selected Felsenstein's model F84 [39,40] for IGSNOD, IGSEXO, IGSGAB, and Felsenstein's model F81 [40,41] for IGSRKP. Then, using the ape package, a set of matrices containing pairwise genetic distances between alleles observed at each locus was computed according to these selected models, and Neighbor-Joining trees with bootstrap values were obtained from these distance matrices to illustrate the data sets (Figure 5).
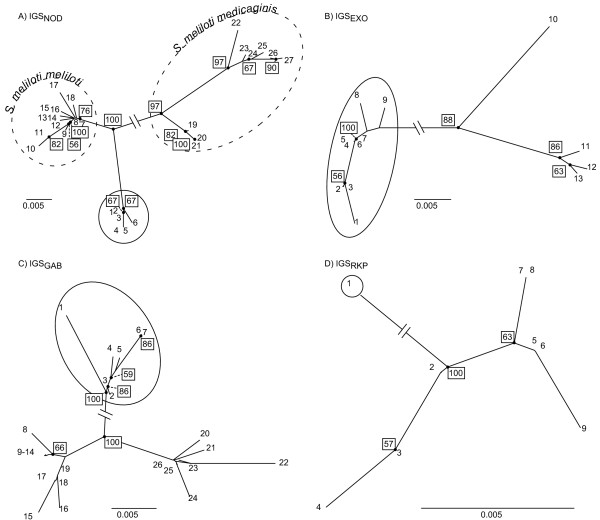
Figure 5.
Neighbor-Joining trees for the representation of the distances among alleles. The alleles belonging to S. medicae isolates are surrounded by a plain-line circle. Only IGSNOD presents alleles found only in S. meliloti bv. meliloti populations and alleles found only in S. meliloti bv. medicaginis. Consequently, for IGSNOD, alleles are also divided according the two biovars of S. meliloti, by broken-line circles. Bootstrap values higher than 50% are given in boxes. Nodes with bootstrap values higher than 50% are indicated by plain circles and in case of possible ambiguity, a broken line links the node to the bootstrap value. The interrupted lines have a length of 0.0986 for IGSNOD, 0.1075 for IGSEXO, 0.0456 for IGSGAB and 0.0421 for IGSRKP.
We applied the multiple DPCoA to this data set, and compared the results to those obtained with STRUCTURE [42,43]. STRUCTURE estimates population structure using genotype data. The basic hypotheses are linkage equilibrium within subpopulations (or possibly weak linkage [44]) and Hardy-Weinberg equilibrium (if the organism under study is not haploid).
Results
Mantel and Rν tests demonstrated that the locus IGSNOD provides a very specific ordination of populations, while the three other markers IGSRKP, IGSEXO and IGSGAB, were significantly congruent (Table 1).
Table 1.
Pairwise correlations among loci with the complete real data set
| Mantel | IGSNOD | IGSEXO | IGSGAB | Rv tests | IGSNOD | IGSEXO | IGSGAB |
| IGSEXO | -0.164 | IGSEXO | 0.232 | ||||
| IGSGAB | -0.173 | 1.000* | IGSGAB | 0.230 | 1.000* | ||
| IGSRKP | -0.164 | 1.000* | 0.999* | IGSRKP | 0.227 | 1.000* | 0.999* |
*Significant correlations with P-values < 0.05.
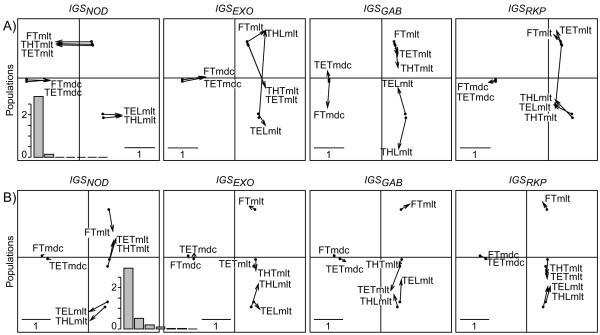
With DPCoA-MCoA (Figure 6), the first axis, which expresses 94% of the diversity among populations, separates the two bacterial species, S. meliloti and S. medicae, while the second axis, with 6% of the diversity among populations, distinguishes the impact of the host plants, M. laciniata and M. truncatula. The DPCoA-STATIS analysis reveals a very similar pattern (Figure 7). Consistently, the STRUCTURE analysis indeed defined two main clusters including respectively S. meliloti and S. medicae, without any trace of admixture between the two species. However, these results are a compromise with the information provided by IGSRKP, IGSGAB, IGSEXO and IGSNOD. Although the four markers effectively delineate the two bacterial species, they express this segregation differently. The DPCoA-MCoA indeed revealed that the segregation between S. meliloti and S. medicae is supported by more than 90% population variation for the three most coherent markers, i.e. IGSRKP, IGSGAB and IGSEXO, while it only concerns a minor part of the population variation observed for IGSNOD. The discrimination between the impact of the two host plants, i.e. M. truncatula and M. laciniata, which appears in axis 2, is the main structure for the IGSNOD marker. The interstructure obtained by using STATIS (Figure 7A), i.e. the eigenanalysis of the Rν matrix, illustrated the special status of IGSNOD.
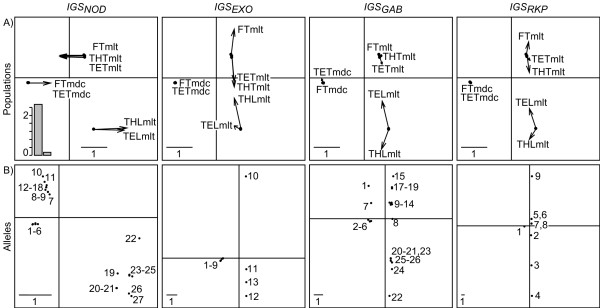
Figure 6.
Application of the DPCoA-MCoA to the real data set. A) Comparison between the patterns of the differences among populations given by the compromise over all loci (black dots, start of the arrows) and the individual analyses (end of the arrows). The special status of IGSNOD is highlighted by horizontal arrows (wrong assignment on the first axis), whereas IGSGAB, IGSRKP and IGSEXO presents vertical arrows (discrepancies from the compromise structure on axis 2 only); B) Location of the alleles. A low (or high) variance in allele points on an axis indicates that the diversity among alleles within populations is lower (or higher) than the diversity among populations, because each axis is normalized for diversity among populations. An eigenvalue barplot is provided in the left-hand corner.
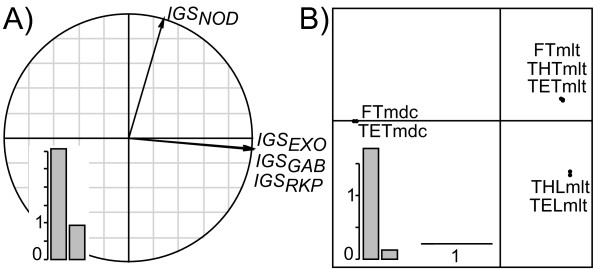
Figure 7.
Application of the DPCoA-STATIS to the real data set. A) The interstructure which displays the eigenanalysis of the Rν matrix, and B) the best compromise. Eigenvalue barplots are provided in boxes. In the interstructure (A), the smaller the angle between two loci, the more similar the inter-population patterns provided by the two loci.
It is noteworthy that based on DPCoA-MCoA, the secondary structure is due to a host-plant effect (e.g. IGSGAB) and/or a geographical origin effect (e.g. IGSEXO) discriminating between French and Tunisian populations of S. meliloti. Interestingly, the effect of geographical distance on the population structure of S. meliloti is not detected by compromise analyses. Because both STATIS and MFA aim at pointing out similarities among loci, these approaches failed at highlighting the secondary structure observed using DPCoA-MCoA (Figure 7B and Figure 8).
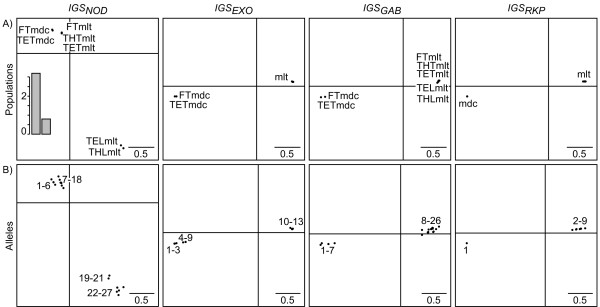
Figure 8.
Application of the DPCoA-MFA to the real data set. A) Patterns of population differences, and B) allele differences per locus. An eigenvalue barplot is provided at the left-hand corner. Only "mlt" (respectively "mdc") is written when no differentiation can be done on the graphs among S. meliloti (respectively S. medicae) populations.
There is a clear relationship between the patterns of population differences and the distribution of allelic diversity (Figure 6B). For instance, the two bacterial species did not share any alleles in common, even for the IGSNOD locus. Furthermore, the populations associated with M. laciniata did not share any alleles with the populations associated with M. truncatula for the IGSNOD locus, resulting in three independent allelic pools belonging respectively to S. medicae and the two biovars of S. meliloti. Furthermore, the distance between the IGSNOD alleles associated with M. laciniata and those associated with M. truncatula is very high, almost as high as the distance which separates S. meliloti and S. medicae on IGSEXO. The particular polymorphism pattern observed for IGSNOD might be explained by both the host-plant selective pressure that acts on nod genes and the events of horizontal transfer that affect the nod gene cluster [34].
Relative effects of distances and frequencies
In order to estimate the relative impacts of allele frequencies and distances in the above results, we applied the DPCoA-MCoA taking into account either sequence divergences without allele frequencies or allele frequencies without sequence divergences (Figure 9). When only sequence divergences are kept, like in the complete analysis, IGSEXO, IGSGAB, and IGSRKP are significantly correlated sharing a strong separation between the species S. medicae and S. meliloti (correlations vary from 0.81 and 0.93 according to Mantel and are superior to 0.999 according to Rν; significance of correlation tests was assessed according to a 0.05 threshold). Regarding the DPCoA-MCoA factorial maps, the population structure is maintained on axis 1, which in that case exhibits 96% of the inter-population diversity. IGSNOD stands out by presenting very distinct alleles according to the host plant. On the second axis, with 4% of the inter-population diversity, the differences between populations according to host plants are maintained for IGSGAB as a secondary structure. Yet, the secondary structures of both IGSRKP and IGSEXO become hardly interpretable. When only the allele frequencies are kept, due to the high differentiation between the two species S. medicae and S. meliloti for all the loci when allele distances are removed, all the pairwise correlations between loci are significant according to the Mantel statistic (correlations greater than 0.83), and all except IGSEXO-IGSNOD (0.61) and IGSRKP-IGSNOD (0.63) correlations according to the Rν statistic. Regarding the DPCoA-MCoA factorial maps, the first axis of all the loci represents the inter-species separation. The difference among populations according to their host plant measured on IGSNOD is relegated to axis 2 representing 12% of the inter-population analysis. Along this axis, all the three other loci IGSEXO, IGSGAB, and IGSRKP distinguish the French population from the Tunisian populations.
Figure 9.
Effects of allele frequencies and distances in thereal data set. We applied the DPCoA-MCoA to A) the data set with allele distances without allele frequencies; B) the data set with allele frequencies, without allele distances. In each of the two cases A) and B), each plot gives a comparison between the patterns of the differences among populations given by the compromise over all loci (black dots, start of the arrows) and the individual analyses (end of the arrows).
The conclusions which can be drawn from these analyses of the effects of distances and frequencies on the inter-population diversity are as follows. In all of the analyses, the most peculiar locus remains IGSNOD. The high separation of populations according to their host plant is due to distinct and distant alleles for IGSNOD and allele distances for IGSGAB. The differences among IGSGAB, IGSRKP, and IGSEXO are due to differentiation patterns among S. meliloti populations. Finally, the distinction between the French and the Tunisian populations mostly relies on allele frequency data.
Discussion
The MDPCoA approach provides a useful tool for: (i) identifying atypical loci by both tests and factorial maps; (ii) describing differences in population structures between groups of congruent loci by factorial maps; (iii) including evolutionary distances among alleles, which is seldom done.
Missing data
In all the analyses we performed, the weight of a population is the number of individuals sampled from this population divided by the total number of individuals sampled. Given that we consider several loci, this definition of the weights supposes that we have identified the allelic composition of each individual for all loci. In case of missing allelic data, i.e. if the allelic content of some individuals is missing for one or several loci, one should define different weight systems depending on the loci. According to the gth locus, the weight of population i is the number of characterized individuals from population i divided by the total number of characterized individuals. This would lead to G different systems of weights, i.e. one per locus. Unfortunately, neither STATIS nor the MCoA nor the MFA can support different population weights. Consequently, one will have to assume a similar set of population weights over loci although some data are missing. To overcome this problem, it may be assumed that the weight of a population is the number of individuals sampled from this population divided by the total number of individuals sampled, whether or not the allelic information for all the loci and for all the individuals is available.
Another case of usual missing data is the lack of nucleotide divergence among alleles. In that case, we suggest fixing the distance among any two different alleles equal to 1, so that the DPCoA is equal to the non-symmetric correspondence analysis [11,45]. Furthermore, the inertia of the allelic points per population in the DPCoA "common space" is then equal to the gene diversity index H, introduced by Nei [28], and the inertia of the population points is equal to the gene diversity among populations defined by Nei [28] in its decomposition of gene diversity. The inertia among population points in the best compromise plot and DPCoA-STATIS is a measure of gene diversity among populations averaged over the G loci, where the weights given to the loci are not simply uniform but set optimal for synthesizing what is common to the loci. This process gives less weight to outliers and reflects the distances among populations as they are seen by the majority of the loci.
Effects of frequencies and distances
The effect of frequencies and distances comprises two components: the effect due to sampling error and the effect due to population structure. The effects of sampling error on the component of nucleotide diversity within and between populations have been studied elsewhere [23,46], and might be the object of further research in the context of the mDPCoA.
The relative effects of frequencies and distances on the analysis of population structure depend on the degree of differentiation among the populations under study. In case of low differentiation, population structure is usually due to variations in allelic frequencies. For instance, differences among French and Tunisian populations of S. meliloti that are highlighted by IGSEXO, IGSGAB and IGSRKP are due to allelic frequencies. Conversely, as the number of alleles shared by the different population decreases, taking into account the information provided by sequence divergence is crucial to efficiently describe their relationships. For instance, the specific inter-population structure of IGSNOD is mainly due to sequence divergence.
Pertinence of the correlation tests
Both correlation tests (Mantel and Rν) can be non-significant for two reasons: either because of an absence of population structure or because the two loci compared reveal different population structures. As highlighted in a previous section, the estimated ϕST parameter and the factorial maps obtained by one of the three versions of the mDPCoA (with MCoA, STATIS or the MFA), can be used to choose among the two alternatives. Concerning the relative interest of the two tests, the Rν test is revealed to be more powerful when applied to our simulated data set, so we advocate its use.
Relative advantages and disadvantages of the three proposed analyses – choice of a method
The three methods are alike in their procedure because they are all based on a compromise. However, they differ in the way the compromise is obtained. With the MCoA, the compromise is built during the definition of the factorial axes. It maximizes the average correlation among the individual analyses and the compromise. With STATIS, the compromise is obtained before going to the core of the multivariate ordination analysis. Here, the compromise maximizes the correlations among the patterns of inter-population diversity provided by the loci. With the MFA, the pieces of information given by the loci are simply added to each other by creating a large table juxtaposing the information on the loci. This last method is the simplest, where pieces of information are simply added. On the other hand, MCoA and STATIS first compare the patterns of inter-population diversity provided by the loci, either for visualizing in a single space the differences among loci or for erasing these differences, and find a best compromise over the loci, respectively.
Unfortunately, the representation of the differences among loci with STATIS is not optimal [15] because STATIS focuses on similarities instead of dissimilarities among loci. Consequently, in comparison to alternative methods, it theoretically lacks an optimal explicability, and an efficient description of the differences in population patterns among loci. The description of the differences among population patterns is thus more precise using MCoA and MFA. Conversely, the main advantage of STATIS over other methods is that it provides a simpler compromise pattern.
The choice among the three methods therefore depends on the goal of the underlying study. If the objective is to obtain the best compromise over the loci, then we advocate the use of DPCOA with STATIS. However, if the objective is to obtain a detailed comparison among the population patterns provided by the G loci, then we encourage the use of the DPCoA with the MCoA.
Complementarity between mDPCoA and other analyses
The mDPCoA could be associated with other tools to study population structure, including the AMOVA, which forms the basis of the DPCoA, Linkage Disequilibrium (LD) statistics, and also recent approaches such as STRUCTURE or CLONAL FRAME.
The AMOVA averages molecular variability over loci to test the existence of differences between populations or groups of populations in terms of both allele frequencies and nucleotide distances among alleles. The Mantel and Rv statistics associated with the mDPCoA use the same information to test the differences between the inter-population structures inferred by several loci.
Both linkage disequilibrium (LD) measures and the mDPCoA aim at assessing whether there is a significant association among the polymorphism patterns observed for different molecular markers. However, LD approaches and mDPCoA differ in several ways. Without discrepancies among the population structures, mDPCoA would fail to detect that different loci evolve independently, even if these are in linkage equilibrium at the population scale. Conversely, in the Sinorhizobium spp. data set, the mDPCoA detected that IGSNOD pattern of population differences was drastically different from the ones obtained with IGSRKP, IGSGAB and IGSEXO, suggesting a horizontal gene transfer of nod genes between S. meliloti bv. meliloti and S. medicae. Because of the differentiation between S. meliloti and S. medicae, LD measures would have failed to detect such a transfer event. Linkage disequilibrium measures and mDPCoA therefore appear as complementary tools to study the influence of sex during the evolution of bacterial lineages.
The mDPCoA is above all a descriptive method, as it does not rely on any assumptions about models of evolution such as linkage equilibrium or selective neutrality. Nevertheless, this analysis pipeline can raise questions that will be investigated using complementary analyses. Thus, demonstrating differences among population structures obtained from different loci raised questions regarding the definition of population boundaries, or the genealogy of both genes and individuals. A consensus population structure could be inferred without any a priori knowledge using STRUCTURE, and its efficiency can be confirmed and illustrated using the correlation tests and the graphical outputs of the mDPCoA. CLONAL FRAME is an explanatory method, estimating clonal relationships and looking for key recombination events with a view of finding the mechanisms implied in microevolution [47]. It can be used to gain insights into the history of an atypical locus. Finally, the detection of selection traces and mechanistic experiments can be of great interest to explain mDPCoA results. These different approaches thus complement the mDPCoA, and conversely, the mDPCoA complements these approaches. For instance, both STRUCTURE and CLONAL FRAME imply working on MLS analyses, and the choice of the finite set of loci used in these analyses may be crucial. Each method can be improved by looking at the results returned by the two others. A joint interpretation of the results of the alternative methods may thus allow a better interpretation of the results and lead to a deeper analysis of particular loci for a better understanding of the data.
Conclusion
All three methods proposed can be used for a better description of inter-population genetic diversity measured over more than one locus. They imply a new reflection on the role of means in measures of diversity: can we work on average information over loci, or do we first need to examine the differences among the patterns of diversity given by the loci? Sometimes, the differences among loci are so high that the compromise obtained by the multivariate analyses will be unstable and the use of averaged information can hamper interpretation. This issue is related to the question raised decades ago: can we build a unique, very synthetic measure of biodiversity, or do we have to make up our mind to define several conflicting measures? As it is based on multivariate analyses, the multiple DPCoA in its three forms can be used to analyze large data sets. It allows a comparison of genetic diversity measured on various loci. It complements existing tools such as AMOVA and linkage disequilibrium measures. It is used here on molecular data because it is in genetics the question of congruence among markers was raised several years ago. We illustrated this procedure using a limited but complex sequence database. The method will have to be tested on other data sets, yet the results are already very promising. Moreover, mDPCoA is potentially more general than we presented here since it can be extended to any data set where pairs of matrices comprise a matrix with abundance or presence/absence and a matrix of dissimilarities. Further applications in ecology could thus be considered, such as the description of inter-community diversity based on both genotypic and phenotypic features.
Abbreviations
AMOVA, Analysis of MOlecular Variance; bv., biovar; DPCoA, Double Principal Coordinate Analysis; FTmdc, Population sampled at Sainte Colombe l'Eglise in France from M. truncatula nodules which include S. medicae isolates; FTmlt, Population sampled at Sainte Colombe l'Eglise in France from M. truncatula nodules which include S. meliloti bv. meliloti isolates; IGS, Intergenic spacers; LD, Linkage disequilibrium; MCoA, Multiple Co-inertia Analysis; mDPCoA, multiple Double Principal Coordinate Analysis; MFA, Multiple Factorial Analysis; MLS, Multilocus Sequencing; PCA, Principal Component Analysis; STATIS, comes from a French expression "structuration des tabeaux à trois indices de la statistique" which means: structuration of the tables characterized by three statistical modes; TELmlt, Population sampled in Tunisia at Enfidha from M. laciniata nodules which include S. meliloti bv. medicaginis isolates; TETmdc, Population sampled in Tunisia at Enfidha from M. truncatula nodules which include S. medicae isolates; TETmlt, Population sampled in Tunisia at Enfidha from M. truncatula nodules which include S. meliloti bv. meliloti isolates; THLmlt, Population sampled in Tunisia at Hadjeb from M. laciniata nodules which include S. meliloti bv. medicaginis isolates; THTmlt, Population sampled in Tunisia at Hadjeb from M. truncatula nodules which include S. meliloti bv. meliloti isolates.
Authors' contributions
SP developed the methodology and applied it to the data. XB performed the simulations and characterized Sinorhizobium populations. He interpreted the results. Both authors contributed equally to the discussion. Both authors read and approved the final draft.
Supplementary Material
Functions in R to perform multiple DPCoA. The file is called "mdpcoa.R". It can be read by the R software which can be downloaded free of charge, and one can refer to the Additional file 2 for explanation on how to use it.
Instructions for performing multiple DPCoA in R. The file is called "Instruction.pdf". It describes in step by step detail how to use R to perform a multiple DPCoA using the real data set in this paper.
Description of the real data set. The complete sampling procedure is given together with a description of within-population diversity.
DNA sequences for IGSNOD. Sequences are in "FASTA" format. The File is named "NOD.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSEXO. Sequences are in "FASTA" format. The File is named "EXO.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSGAB. Sequences are in "FASTA" format. The File is named "GAB.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSRKP. Sequences are in "FASTA" format. The File is named "RKP.aa". See Additional file 2 for explanation on how to use this file.
Acknowledgments
Acknowledgements
The authors are grateful to Pr. I Olivieri, Pr. JPW Young and two anonymous reviewers for their useful comments about this study. We also thank R. Lower, and the American Journal Experts who helped us to improve the quality of this manuscript. This paper takes place in a research project on "Biodiversity, perception and use" funded by the French Institute of Biodiversity. Within this more general context, we develop and discuss methodologies for measuring biodiversity on multi-marker data sets at various scales, from individuals' gene loci to species' functional traits.
Contributor Information
Sandrine Pavoine, Email: pavoine@mnhn.fr.
Xavier Bailly, Email: xb502@york.ac.uk.
References
- Cooper JE, Feil EJ. Multilocus sequence typing: what is resolved? Trends in Microbiology. 2004;12:373–377. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Spratt BG. The impact of homologous recombination on the generation of diversity in bacteria. Journal of Theoretical Biology. 2006;239:210–209. doi: 10.1016/j.jtbi.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1968–1973. doi: 10.1073/pnas.0406993102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Emerging technologies in DNA sequencing. Genome Research. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- Moazami-Goudarzi K, Laloë D. Is a multivariate consensus representation of genetic relationships among populations always meaningful? Genetics. 2002;162:473–484. doi: 10.1093/genetics/162.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biology. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Torpdahl M, Didelot X, Conrad DF, Wilson DJ, Achtman M. Mismatch induced speciation in Salmonella: model and data. Philosophical Transactions of the Royal Society of London Series B - Biolog. 2006;361:2045–2053. doi: 10.1098/rstb.2006.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly X, Olivieri I, De Mita S, Cleyet-Marel JC, Béna G. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Molecular Ecology. 2006;15:2719–2734. doi: 10.1111/j.1365-294X.2006.02969.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Megraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Escoufier Y. Le traitement des variables vectorielles. Biometrics. 1973;29:750–760. doi: 10.2307/2529140. [DOI] [Google Scholar]
- Pavoine S, Dufour AB, Chessel D. From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. Journal of Theoretical Biology. 2004;228:523–537. doi: 10.1016/j.jtbi.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessel D, Hanafi M. Analyses de la co-inertie de K nuages de points. Revue de Statistique Appliquée. 1996 http://www.numdam.org/item?id=RSA_1996__44_2_35_0 [Google Scholar]
- Lavit C, Escoufier Y, Sabatier R, Traissac P. The ACT (Statis method) Computational Statistics and Data Analysis. 1994;18:97–119. doi: 10.1016/0167-9473(94)90134-1. [DOI] [Google Scholar]
- Escofier B, Pagès J. Multiple factor analysis: results of a three-year utilization. In: Coppi R and Bolasco S, editor. Multiway data analysis. , Elsevier Science Publishers B.V., North-Holland; 1989. pp. 277–285. [Google Scholar]
- Chessel D, Dufour AB, Thioulouse. J. The ade4 package -I- One-table methods. R News. 2004;4:5–10. http://cran.r-project.org/doc/Rnews/Rnews_2004-1.pdf [Google Scholar]
- Paradis E, Strimmer K, Claude J, Jobb G, Opgen-Rhein R, Dutheil J, Noel Y, Bolker B. ape: Analyses of Phylogenetics and Evolution. , R package version 1.7; 2005. [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. doi: 10.2307/1390807. [DOI] [Google Scholar]
- Gower JC. Euclidean distance geometry. Mathematical Scientist. 1982;7:1–14. [Google Scholar]
- Lingoes JC. Some boundary conditions for a monotone analysis of symmetric matrices. Psychometrika. 1971;36:195–203. doi: 10.1007/BF02291398. [DOI] [Google Scholar]
- Cailliez F. The analytic solution of the additive constant problem. Psychometrika. 1983;48:305–310. doi: 10.1007/BF02294026. [DOI] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CR. Diversity and dissimilarity coefficients: a unified approach. Theoretical Population Biology. 1982;21:24–43. doi: 10.1016/0040-5809(82)90004-1. [DOI] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavoine S, Dolédec S. The apportionment of quadratic entropy: a useful alternative for partitioning diversity in ecological data. Environmental and Ecological Statistics. 2005;12:125–138. doi: 10.1007/s10651-005-1037-2. [DOI] [Google Scholar]
- Rao CR. Rao's axiomatization of diversity measures. In: Kotz S and Johnson NL, editor. Encyclopedia of Statistical Sciences. New York, Wiley and Sons; 1986. pp. 614–617. [Google Scholar]
- Nei M. Analysis of gene diversity in subdivised populations. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY, USA, Columbia University Press; 1987. [Google Scholar]
- Laval G, Excoffier L. SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics. 2004;12:2485–2487. doi: 10.1093/bioinformatics/bth264. [DOI] [PubMed] [Google Scholar]
- Kimura M. Stepping Stone model of population. Annual Report of the National Institute of Genetics. 1953;3:62–63. [Google Scholar]
- Jukes T, Cantor C. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York, Academic press; 1969. pp. 21–132. [Google Scholar]
- Charlesworth D, Mable BK, Schierup MH, Bartolomé C, Awadalla P. Diversity and Linkage of Genes in the Self-Incompatibility Gene Family in Arabidopsis lyrata. Genetics. 2003;164:1519–1535. doi: 10.1093/genetics/164.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly X, Olivieri I, Brunel B, Cleyet-Marel JC, Béna G. Horizontal gene transfer and homologous recombination drive the evolution of the nitrogen-fixing symbionts of Medicago species. Journal of Bacteriology. 2007;189:5223–5236. doi: 10.1128/JB.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bena G, Lyet A, Huguet T, Olivieri I. Medicago - Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago. Journal of Evolutionary Biology. 2005;18:1547–1558. doi: 10.1111/j.1420-9101.2005.00952.x. [DOI] [PubMed] [Google Scholar]
- Villegas MDC, Rome S, Maure L, Domergue O, Gardan L, Bailly X, Cleyet-Marel JC, Brunel B. Nitrogen-fixing sinorhizobia with Medicago laciniata constitute a novel biovar (bv. medicaginis) of S. meliloti. Systematic and Applied Microbiology. 2006;29:526–538. doi: 10.1016/j.syapm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Barran LR, Bromfield ES, Brown DC. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti - Medicago laciniata symbiosis. Canadian Journal of Microbiology. 2002;48:765–771. doi: 10.1139/w02-072. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Felsenstein J, Churchill GA. A Hidden Markov model approach to variation among sites in rate of evolution. Molecular Biology and Evolution. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. http://mbe.oxfordjournals.org/cgi/content/abstract/13/1/93 [DOI] [PubMed] [Google Scholar]
- McGuire G, Prentice MJ, Wright F. Improved error bounds for genetic distances from DNA sequences. Biometrics. 1999;55:1064–1070. doi: 10.1111/j.0006-341X.1999.01064.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;Published article online doi: 10.1111/j.1471-8286.2007.01758.x doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure: Extensions to linked loci and correlated allele frequences. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro N, D'Ambra L. L'analyse non symétrique des correspondances. In: Diday E, Jambu M, Lebart L, Pages J and Tomassone R, editor. Data Analysis and Informatics, III. North-Holland, Elsevier; 1984. pp. 433–446. [Google Scholar]
- Lynch M, Crease TJ. The analysis of population survey data on DNA sequence variation. Molecular Biology and Evolution. 1990;7:377–394. doi: 10.1093/oxfordjournals.molbev.a040607. http://mbe.oxfordjournals.org/cgi/content/abstract/7/4/377 [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference on bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functions in R to perform multiple DPCoA. The file is called "mdpcoa.R". It can be read by the R software which can be downloaded free of charge, and one can refer to the Additional file 2 for explanation on how to use it.
Instructions for performing multiple DPCoA in R. The file is called "Instruction.pdf". It describes in step by step detail how to use R to perform a multiple DPCoA using the real data set in this paper.
Description of the real data set. The complete sampling procedure is given together with a description of within-population diversity.
DNA sequences for IGSNOD. Sequences are in "FASTA" format. The File is named "NOD.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSEXO. Sequences are in "FASTA" format. The File is named "EXO.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSGAB. Sequences are in "FASTA" format. The File is named "GAB.aa". See Additional file 2 for explanation on how to use this file.
DNA sequences for IGSRKP. Sequences are in "FASTA" format. The File is named "RKP.aa". See Additional file 2 for explanation on how to use this file.