Abstract
The familial breast–ovarian tumor suppressor gene product BRCA1 was found to be a component of the RNA polymerase II holoenzyme by several criteria. BRCA1 was found to copurify with the holoenzyme over multiple chromatographic steps. Other tested transcription activators that could potentially contact the holoenzyme were not stably associated with the holoenzyme as determined by copurification. Antibody specific for the holoenzyme component hSRB7 specifically purifies BRCA1. Immunopurification of BRCA1 complexes also specifically purifies transcriptionally active RNA polymerase II and transcription factors TFIIF, TFIIE, and TFIIH. Moreover, a BRCA1 domain, which is deleted in about 90% of clinically relevant mutations, participates in binding to the holoenzyme complex in cells. These data are consistent with recent data identifying transcription activation domains in the BRCA1 protein and link the BRCA1 tumor suppressor protein with the transcription process as a holoenzyme-bound protein.
BRCA1 is a tumor suppressor gene that is mutated in a significant fraction of cases of inherited breast and ovarian cancer. Approximately 3% of breast cancer is attributable to inherited mutations in BRCA1. Indeed, in ∼50% of families with an abnormally high incidence of breast cancer through multiple generations, the offending mutation is in the BRCA1 gene (1–3). The BRCA1 product is likely to have tumor suppression function, since tumors arising in members of BRCA1-linked families show loss of heterozygosity at the BRCA1 locus, with retention of the mutant/disease-predisposing allele (4, 5).
The BRCA1 gene encodes a 1,863-amino acid protein without extensive homology to other proteins (1, 2). The primary sequence is noteworthy for a RING-finger motif and an acidic carboxyl terminus (1), both of which are characteristics of certain transcription factors. The BRCA1 7.8-kb mRNA is observed in many tissues, with expression highest in testis and thymus (1). About 90% of the mutations observed in the BRCA1 gene result in truncations, and the remainder of clinically relevant mutations are individual missense abnormalities that are scattered along the entire coding unit (3).
A defined carboxyl-terminal segment of BRCA1 can activate transcription when fused to the DNA binding domain of GAL4 (6, 7). These fusion proteins activated transcription from promoters containing a GAL4 binding site. Importantly, fusion proteins bearing clinically relevant point mutations were inactive in this assay, implying, at a minimum, that the transcription assay is a faithful monitor of the native structure of a segment of the protein. Although there are other interpretations, these data have licensed the speculation that BRCA1 is, at least in part, a transcription factor. Whatever the significance of the transactivation potential of its carboxyl-terminal region, there is now evidence pointing to a role for BRCA1 in the control of DNA repair and genome stability (8). Hence, if it proves to have genuine transcription regulation function, it will be interesting to determine whether these two functions relate to one another and, if so, how.
Mammalian RNA polymerase II (pol II) exists in two forms: one now known as “core” polymerase containing 10 to 12 subunits and a mass of about 500 kDa and a second form known as the “holoenzyme” containing multiple subunits and a mass in excess of a megadalton. SRB proteins are key components of the holoenzyme and were discovered in a yeast genetic screen as suppressors of RNA polymerase B mutations. Nine different SRB proteins bind to the carboxyl-terminal domain (CTD) of yeast pol II and are only found in the pol II holoenzyme (9). The yeast holoenzyme has been well characterized and was found to contain not only pol II and the SRB proteins but also the basal transcription factors TFIIB, TFIIF, and TFIIH. The transcriptional coactivator GAL11 has been detected in the complex (10) along with the SWI–SNF chromatin remodeling complex (11). The available data indicate that the yeast holoenzyme is responsible for all mRNA transcription initiation in a cell, since a temperature-sensitive mutation of one SRB protein rapidly eliminated transcription at the restrictive temperature (12).
The mammalian counterpart of the yeast holoenzyme has only recently been described. It was found to contain several of the basal transcription factors and three SRB homologues (13–15). The human homologue of yeast SRB7 was cloned and shown to function in yeast cells by complementing partial deletion mutants of SRB7 (14). The presence of one or more specific SRBs in the mammalian holoenzyme complex was viewed as significant since antibody to hSRB7 was used to follow the purification of enzymatically active complex from calf thymus extracts.
Yeast SRB10 and SRB11 are a cyclin–kinase pair (16), and the human proteins cdk8 and cycC share sequence homology with these two proteins, respectively (17, 18). The latter two proteins were also detected in the mammalian holoenzyme (15). The basal transcription factors TFIIE and TFIIH were also shown to be in the complex by coimmunoprecipitation, although during large-volume biochemical purification, much of the TFIIE and TFIIH was lost (14), suggesting that the mammalian holoenzyme is experimentally less stable than its yeast counterpart.
To address this experimental problem, we have developed a new purification strategy for the mammalian holoenzyme and used it to search for specific transcription factors in the holoenzyme complex. Unlike sequence-specific DNA-binding transcription activators that were analyzed, wild-type BRCA1 protein regularly copurified with the holoenzyme. Although all of the evidence on BRCA1 and transcription has, so far, been obtained by reconstruction experiments that involved the ectopic production of truncated BRCA1 polypeptides fused to a foreign DNA binding domain, the results presented herein imply a physiological role for intact full-length BRCA1 in the regulation of transcription of at least some genes.
MATERIALS AND METHODS
Purification of pol II Holoenzyme.
HeLa whole-cell extracts were prepared by a modification of the standard method (19) in which all chloride anions were replaced by acetate. The extract was dialyzed into buffer H (20 mM Tris-OAc, pH 7.9/1 mM EDTA/5% glycerol/1 mM dithiothreitol/1 mM phenylmethylsulfonyl fluoride) plus 0.15 M KOAc and applied to a Bio-Rex 70 column at a ratio of 1 ml of matrix per 15–20 mg of protein. Protein fractions were step-eluted in 0.3 M KOac, 0.6 M KOAc, and 1.5 M KOAc, all in buffer H. Part of the 0.6 M step (6 ml, 20 mg) was layered on a 29-ml sucrose step gradient (10–60% sucrose in 20 mM Tris-OAc, pH 7.9/0.2 M KOac/1 mM EDTA/0.1% Nonidet P-40) in SW28 centrifuge tubes (Beckman). After centrifugation at 25,000 rpm at 4°C for 16 h, the bottom of the tube was punctured, and 1-ml fractions were collected. Holoenzyme-containing fractions detected by Western blotting were passed through a G-50 column for salt exchange into 0.15 M KOAc/20 mM Tris-OAc, pH 7.9/5 mM imidazole/20% sucrose. Sample was applied to 1 ml of Ni-nitrilotriacetate matrix (Qiagen, Chatsworth, CA) by batch binding for 1 h at 4°C, and, after extensive washing of the matrix, a 12-ml linear gradient from 5 mM to 130 mM imidazole was used to elute holoenzyme.
Immunoprecipitation.
Protein from the Bio-Rex 0.6 M peak was immunoprecipitated with the indicated antibody. Binding reactions were rotated overnight at 4°C in buffer H/0.12 M KOAc/0.1% Nonidet P-40/0.1 mM dithiothreitol/BSA (0.2 mg/ml)/1 mM phenylmethylsulfonyl fluoride in the presence of protein extract, antibody, and protein A beads. The supernatant was removed, and the protein A beads were then washed three or four times in 40 vol of 20 mM Tris-OAc, pH 7.9/0.12 M KOAc/6 mM Mg(OAc)2/0.1% Nonidet P-40/0.1 mM dithiothreitol/BSA (0.2 mg/ml). For Western blot analysis, samples were subjected to electrophoresis in either 4% or 12.5% SDS/PAGE gels and immunoblotted.
In vitro transcription assays from immunoprecipitates were performed as in ref. 14 except that the salt was 0.1 M KOAc instead of 0.06 M KCl.
Transfection and Immunoprecipitation.
The 293T cells were transfected with 10 or 20 μg of plasmid DNA expressing BRCA1 with the hemagglutinin (HA) tag fused to the amino terminus (8). Wild-type protein (1,863 amino acids plus the tag) or truncated mutant (1,852 amino acids plus the tag) were detected by pulse labeling with [35S]methionine and immunoprecipitation using anti-HA antibody. For analysis of holoenzyme complexes, lysates from transfected cells were prepared in 0.5 M KOAc/0.25% Nonidet P-40/50 mM Tris-OAc, pH 7.9/10 mM EDTA/1 mM phenylmethylsulfonyl fluoride/10 mM β-glycerophosphate/5 mM NaF. Lysates were either subjected to SDS/PAGE for evaluation of steady-state levels or were immunoprecipitated with affinity-purified anti-SRB7 antibody and then subjected to SDS/PAGE. The amount of recovered protein was analyzed by immunoblotting and probing with anti-HA tag antibody.
RESULTS
Biochemical Purification of the Holoenzyme and BRCA1.
HeLa whole-cell extract was applied to a Bio-Rex 70 column, and protein was eluted in increasing concentrations of potassium acetate. Virtually all of the BRCA1 p220 polypeptide coeluted with half of the polymerase and virtually all of the human SRB10 homologue cdk8 (Fig. 1a). From the results of multiple repetitions of this experiment, it appeared that ≈70% of the extract protein did not bind to the column, 10% was in the 0.3 M peak, 12% was in the 0.6 M protein peak, and 5% was in the 1.5 M peak. There is no quantitative measure of holoenzyme recovery, but, from Western blotting and measurements of pol II transcription activity, it appears that the recovery of holoenzyme was nearly 100% (data not shown). Thus, the elution of holoenzyme in the 0.6 M protein peak from this ion-exchange column represents an ≈8-fold purification.
Figure 1.
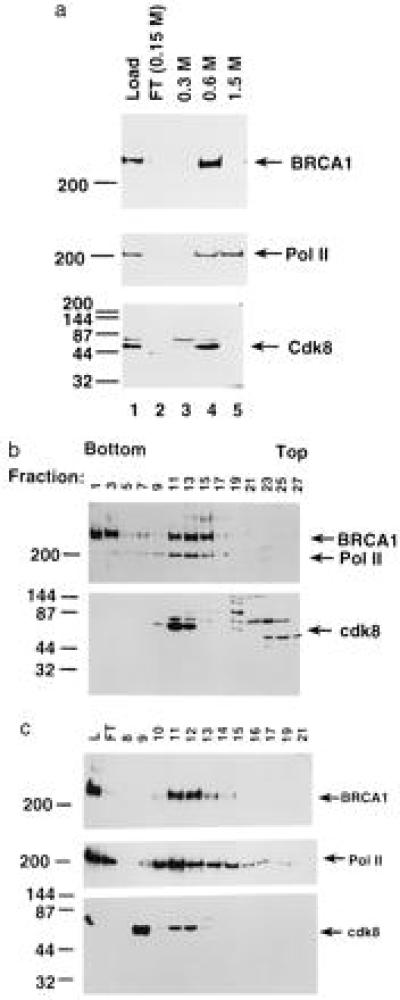
BRCA1 copurifies with the holoenzyme complex. (a) HeLa whole-cell extract was chromatographed on Bio-Rex 70 matrix and step-eluted at 0.3 M, 0.6 M, and 1.5 M KOAc. Protein samples from each fraction were subjected to SDS/PAGE, and blots were probed with BRCA1, pol II, and cdk8 antibodies. (b) The Bio-Rex 70 0.6 M fraction was subjected to centrifugation through a 10–60% sucrose gradient in 0.2 M acetate salt and 0.1% Nonidet P-40. After centrifugation, samples were collected, analyzed for total protein content, and Western-blotted. Blots were probed for BRCA1, pol II large subunit, and cdk8. BRCA1 pool A includes fractions 10–15, and BRCA1 pool B includes fractions 1–3. (c) Sucrose sedimentation pool A was subjected to metal chelate chromatography. Fractions were eluted with a linear 5–130 mM gradient of imidazole. The indicated fractions were subjected to immunoblot analysis and probed with antibodies specific to BRCA1 (Top), pol II large subunit (Middle), and cdk8 (Bottom).
The 0.6 M Bio-Rex 70 column protein fraction was centrifuged through a sucrose gradient, and a peak containing BRCA1, pol II, and cdk8 was identified by Western blotting. This peak separated from the bulk of the protein was applied to the gradient (Fig. 1b). These three polypeptides appeared in two peaks, a major peak in fractions 11–15 (peak A) and a second peak (peak B) centering on fraction 3. The latter had an extremely high sedimentation rate and separated from >99% of the applied protein. Both of these fractions probably have markedly elevated molecular weights, since a 270-kDa marker protein sedimented near the top of the gradient (i.e., in fractions 23–27; data not shown). The protein in peak A contained the highest concentrations of polymerase and cdk8 and about 50% of the input BRCA1. Minimal amounts of pol II and cdk8 were detected in peak B upon prolonged exposure. Since peak B was relatively low in polymerase and cdk8, this fraction may represent a BRCA1-containing complex distinct from the pol II holoenzyme. Further characterization of the protein in peak B was not pursued. The purification of the holoenzyme in peak A was about 5-fold, since virtually all of the holoenzyme was present along with ≈20% of the recovered protein in these fractions.
The material from sucrose gradient peak A was then subjected to metal chelate chromatography (Fig. 1c), and BRCA1 and cdk8 again coeluted with the polymerase. Almost all of the BRCA1 bound to the column, and, upon exposure to a linear gradient of imidazole, BRCA1 was eluted as a peak in fractions 11 and 12. About 50% of pol II bound to the metal chelate matrix, and the eluted protein, although present in a greater number of fractions, also peaked in fractions 11 and 12. All of the cdk8 bound to the Ni matrix and eluted in two separable peaks, one of which coincided with the BRCA1 and pol II peak (Fig. 1c). The resolution of two peaks of cdk8 indicates that the gradient of imidazole resolved different complexes. None of these proteins contains a run of histidines. Thus, it is possible that their binding and elution are determined by strong interactions with yet other polypeptides in the holoenzyme complex that contain oligohistidine stretches. Clearly, BRCA1, pol II, and cdk8 behave as a relatively stable complex, as determined by cosedimentation in sucrose gradients and coelution during metal chelate chromatography. Fractions 10–13 from the metal chelate column represent about 9% of the recovered protein in that experiment, suggesting an 11-fold purification of the holoenzyme during that step. Thus, after the three purification steps, there was ≈400-fold purification of the holoenzyme. Since there is, as yet, no objective measure of holoenzyme activity and since each of its components may exist also in the free state, the calculated 400-fold purification is meant as an estimate. Several other conventional chromatographic steps were attempted during this phase of the work, and all resulted in a complete loss of detectable pol II activity, suggesting either its irreversible binding to the column matrix or extremely broad elution resulting in subthreshold detection.
BRCA1 Copurification with the Holoenzyme Is Distinct from Other Transcriptional Activators.
The purification scheme was analyzed for other transcriptional regulators. Bio-Rex 70 and sucrose gradient protein fractions were analyzed by Western blotting, in which specific antibodies against a panel of transcription factors served as probes (Fig. 2). The 65-kDa YY1 polypeptide (20) did concentrate in the 0.6 M fraction, as did the holoenzyme. However, upon sucrose gradient sedimentation, it failed to cosediment with either of the two holoenzyme peaks. Although YY1 was found to be a component of the holoenzyme by a different purification strategy (15), it did not associate with this complex when the latter was isolated by this new method. The 120-kDa initiator motif binding protein, TFII-I (21), was observed in multiple Bio-Rex 70 fractions, suggesting the existence of multiple forms of the factor. Less than 25% of it was in the Bio-Rex 0.6 M fraction, and when this fraction was analyzed by sucrose gradient sedimentation, TFII-I separated from the BRCA1–pol II complex. Ninety-five percent of the RelA subunit of NF-κB was in the flow through of the Bio-Rex 70 column and, thus, separated from the holoenzyme. The amount of p65 Rel remaining in the 0.6 M fraction was too low to be detected after sucrose gradient sedimentation (data not shown). The DNA-binding transcriptional activator RBP-Jκ (22) similarly does not copurify with the holoenzyme. The upper band of the doublet is the RBP-Jκ, and it is found in the Bio-Rex 70 0.6 M fraction. However, upon sucrose gradient sedimentation analysis, it was detected in the low molecular weight fractions (Fig. 2).
Figure 2.
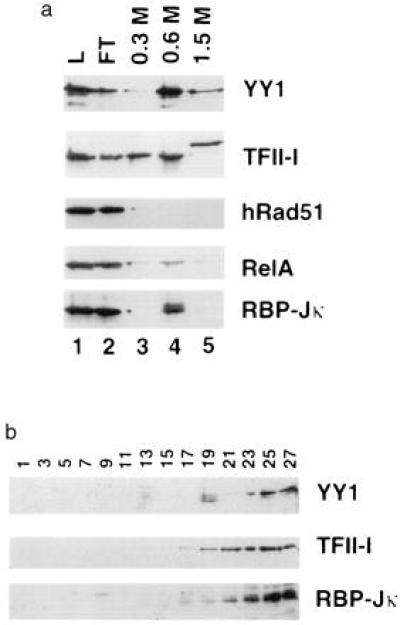
Differential purification of other transcription regulators. (a) The protein fractions from the Bio-Rex 70 chromatographic step were analyzed for the presence of YY1, TFII-I, the RelA subunit of NF-κB, RBP-Jκ, and hRad51. In each case, the indicated bands were the only significant bands of their type observed in the whole blot and migrated at the appropriate position relative to molecular weight markers. (b) Fractions from the sedimentation analysis of the 0.6 M Bio-Rex fraction were analyzed for the presence of YY1, TFII-I, and RBP-Jκ. The protein samples were analyzed for the presence of the RelA subunit of NF-κB and for hRad51, but they were not detected in these fractions.
It has been reported that hRad51 associates with BRCA1 (8) and that hRad51 is a component of the pol II holoenzyme (15). As can be seen in the Western blot analysis of Fig. 2, hRad51 was present only in the Bio-Rex 70 flow-through fraction and was undetectable after gradient sedimentation analysis (data not shown). Thus, hRad51 is not a component of this holoenzyme preparation. As in the case of YY1, hRad51 has been shown to be a holoenzyme component when purified with anti-TFIIF monoclonal antibody followed by several chromatographic steps (15). It is possible that the difference in preparation methods is responsible for the different results observed herein and in the past (15).
hRad51 associates with BRCA1 in vitro and in vivo (8), and comparison of Figs. 1 and 2 shows that the two major pools of BRCA1 observed in these experiments did not copurify with hRad51. The association of BRCA1 and hRad51 appears to be cell cycle dependent, with the resulting complexes being most abundant during S phase (8). Hence, since the extracts studied herein were from asynchronous cells, it is possible that these complexes were insufficiently abundant to be detected because the cells that were extracted were insufficiently enriched for those in S phase. Alternatively, BRCA1–Rad51 complexes may not be stable enough to be detected by the purification methodology used and/or these complexes contain but a small fraction of the total quantity of these proteins present in a cell and were, thus, not apparent in these experiments.
Thus, these data indicate that BRCA1, unlike a series of four transcription regulating proteins, specifically copurified with the pol II holoenzyme.
Coimmunopurification of BRCA1 and Holoenzyme.
Human SRB7 (hSRB7) is a component of the holoenzyme, and antibodies to it can specifically coprecipitate the holoenzyme (14). Immunoprecipitation of the Bio-Rex 0.6 M protein fraction and Western blot analysis of the precipitated proteins revealed the presence of BRCA1 in hSRB7 immunoprecipitates (Fig. 3a). Blocking the anti-hSRB7 immunoprecipitation with the peptide against which the antibody was raised resulted in the loss of the BRCA1 band on immunoblots, while a control peptide was inactive in this regard (Fig. 3b). This demonstrates that the coprecipitation of BRCA1 by affinity-purified hSRB7 antibody was specific. Pol CTD antibody (23) served as another negative control. This antibody binds to the CTD and is known to disrupt the holoenzyme complex (24). In the experiment shown, it failed to coprecipitate BRCA1, unlike the hSRB7 antibody. As yet another negative control, anti-E1A antibody (mAbM73) also failed to coprecipitate BRCA1 (Fig. 3a, lanes 2 and 3).
Figure 3.
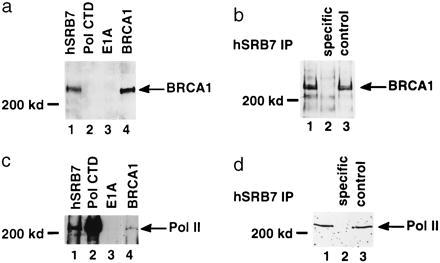
BRCA1 coimmunopurifies with the holoenzyme. The 0.6 M column fraction from the Bio-Rex 70 column was immunoprecipitated with a panel of affinity-purified polyclonal or monoclonal antibodies indicated above each lane in a and c. The pol II antibody (23) is directed at the CTD of the large subunit and disrupts the SRB interaction with the core polymerase (24). In a, the immunoprecipitates were developed with anti-BRCA1 antibody (mAb MS110). In b, the hSRB7 immunoprecipitation was repeated but was competed with specific antigenic peptide (lane 2) or with control peptide (lane 3) and probed with mAb MS110. In c, proteins purified by the same immunoprecipitating antibodies as in a were probed with antibody to the large subunit of pol II, and the pol II polypeptide is indicated. In d, the anti-hSRB7 immunoprecipitation of pol II was performed in the absence and presence (lane 2) of antigenic peptide and the blot was probed with pol II antibody.
Pol II large subunit was coimmunoprecipitated by antibodies raised against hSRB7 and BRCA1 (Fig. 3c). Coimmunoprecipitation of the pol II large subunit by hSRB7 antibody was blocked by preincubation with the relevant antigenic peptide but not a control peptide (Fig. 3d). The yields of pol II in the three different immunoprecipitates shown in Fig. 3c were all different. This difference in yield may reflect the different avidities of each antibody and/or differences in stoichiometry of a given subunit. For example, although BRCA1 appears to be a component of the holoenzyme, it may only be present in a subpopulation of the holoenzyme.
BRCA1 Is Associated with Transcriptionally Active pol II, TFIIF, TFIIE, and TFIIH.
The transcription activity of BRCA1-containing holoenzyme complexes was evaluated by assaying for the presence of pol II activity in anti-BRCA1 immunoprecipitates. The Bio-Rex column fractions enriched in holoenzyme were incubated with a panel of antibodies in the presence of protein A-agarose. The washed immunoprecipitates were then added to transcription reaction mixtures that contained each of the basal transcription factors (TBP, TFIIB, TFIIF, TFIIE, TFIIH) and a G-less cassette template that had been relaxed by topoisomerase I. No pol II was added to this mixture. As can be seen in Fig. 4a, antibodies against hSRB7, pol CTD, and BRCA1 coprecipitated polymerase activity. The BRCA1 monoclonal antibodies used were raised against three distinct antigenic domains, mAb SG11 against a carboxyl-terminal peptide, mAb AP16 against a carboxyl-terminal glutathione S-transferase fusion protein, and mAb MS13 against an amino-terminal glutathione S-transferase fusion protein (25). As shown in Fig. 4a, three antibodies to different epitopes of BRCA1 reproducibly coimmunoprecipitated polymerase activity.
Figure 4.
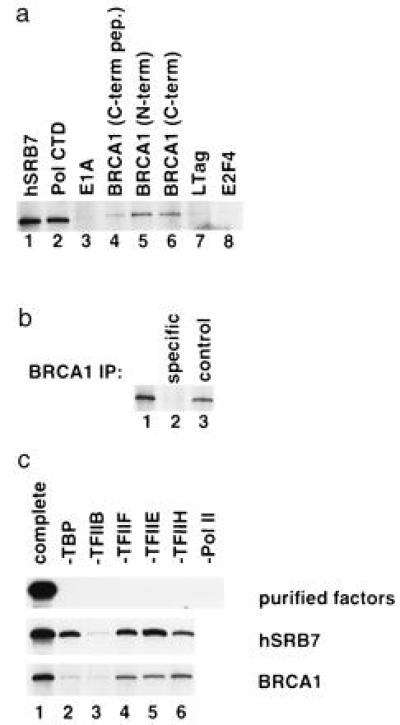
Immunoisolation of BRCA1 copurifies pol II and basal transcription factors. The Bio-Rex 70 0.6 M fraction (20 μg) was immunoprecipitated with the indicated antibodies, and transcription assays were performed on the washed beads. (a) Transcription assays contained each of the basal transcription factors except pol II. Antibodies to hSRB7 (lane 1), pol CTD (lane 2), adenovirus E1A protein (lane 3), BRCA1 carboxyl-terminal peptide (mAb SG11; lane 4), BRCA1 amino-terminal fragment (mAb MS13; lane 5), BRCA1 carboxyl-terminal fragment (mAb AP16; lane 6), simian virus 40 large tumor antigen (lane 7), and transcription factor E2F4 (lane 8) were used. (b) Transcription assays as in a were repeated with mAb SG11 (anti-BRCA1), and antibody binding was competed with 20 μg of antigenic peptide (lane 2) or 20 μg of control peptide (lane 3). (c) The basal factor composition of the proteins present in immunoprecipitates was assayed by supplying all basal factors except the indicated factor noted above each lane. The purified factors were analyzed (Top), and the film exposure time is the same time as in subsequent panels. When a band appears in a given lane, it means that the immunoprecipitate contains the basal activity identified above that lane. Polymerase was not separately added to transcription reactions from immunoprecipitates. Thus, for any transcription to occur, an immunoprecipitate must contain pol II as well as the indicated basal factor.
As controls, antibodies to E1A, the simian virus 40 large tumor antigen, or transcription factor E2F4 failed to coprecipitate polymerase activity. Moreover, mAb SG11 failed to coprecipitate polymerase activity when preincubated with its cognate epitope containing peptide (Fig. 4b). This demonstrates the specificity of the anti-BRCA1 coprecipitation of pol II.
A hallmark of the pol II holoenzyme is that it contains not only polymerase but also certain other basal transcription factors. In one report, TFIIE and TFIIH were associated with the complex (14). In other reports, TFIIF, TFIIE, and TFIIH (15) or all of the basal factors (13) were associated with the holoenzyme. It is possible that, in vivo, the holoenzyme associates with all of these basal factors, but during biochemical purification, some of these activities are lost.
We, thus, asked which of these basal factors, if any, copurified with BRCA1. In preparation for that test, highly purified pol II and five purified basal factors were mixed and found to generate significant RNA synthesis on a suitable template (Fig. 4c Top). In addition, the presence and importance of each of the basal factors in the mixture was assayed by leaving that factor out of the transcription reaction and asking whether RNA synthesis had occurred. Hence, five of the six factors were included in each subsequent reaction mixture. Thus, in lane 2, every factor except TBP was present. In lane 3, all factors except TFIIB were included. In the last lane, pol II was omitted. The results show that the purified basal factors were relatively free of cross-contamination, since only with all present was there effective RNA synthesis.
These reagents were then used to assay for the presence of a given basal factor in the hSRB7 and BRCA1 immunoprecipitates. In these latter two cases, purified pol II was omitted in all lanes as well as the indicated factor. Thus, for transcription activity to be present, the immunoprecipitate must contain substantial pol II and the tested basal factor. Moreover, data were from gels autoradiographed for the same time to insure comparability of results.
An hSRB7 immunoprecipitate (Fig. 4c Middle) revealed transcription activity in the absence of added TFIIF, TFIIE, TFIIH, and TFIID, indicating that these factors were likely present in the hSRB7 immune complexes. TFIIB activity was also present in hSRB7 immunoprecipitates, but at low level.
BRCA1 antibodies to three different epitopes coprecipitated transcription activity when TFIIF, TFIIE, and TFIIH were omitted, implying that these factors were present in the immune complex (Fig. 4c Bottom and data not shown). Thus, like hSRB7 in the holoenzyme complex, BRCA1 appears to interact with RNA pol II and the basal transcription factors TFIIF, TFIIE, and TFIIH.
Association of Wild-Type BRCA1 with Holoenzyme in Cells.
Whether a clinically significant BRCA1 mutation affects holoenzyme binding was also tested. Of the mutations in BRCA1 observed in affected families, 90% result in carboxyl-terminal truncations of various sizes; the smallest deletion being 11 residues (3). Epitope-tagged BRCA1 bearing a mutation that truncates the 1,863-amino acid protein by 11 residues from the carboxyl terminus was synthesized in 293T cells. Cell lysates were prepared from the transfected cells, and holoenzyme was precipitated with antibody against hSRB7, as described in Fig. 3. The mutant BRCA1 species coprecipitated with the holoenzyme less efficiently than the wild type (Fig. 5a). As a measure of the efficiency of transfection, wild-type and mutant BRCA1 were each synthesized at the same rate as determined by pulse labeling (Fig. 5b). In comparing wild-type and mutant BRCA1 protein steady-state levels, Fig. 5c, lanes 1 and 4, revealed similar levels of wild-type and mutant HA-BRCA1 in the lysates, and yet only the wild-type associated significantly with the hSRB7-containing complex (Fig. 5, compare a, lanes 1 and 4, with c, lanes 1 and 4). These results suggest that in cells, BRCA1 associates with the pol II holoenzyme and that the carboxyl-terminal 11 amino acid residues of BRCA1 are important for holoenzyme binding.
Figure 5.
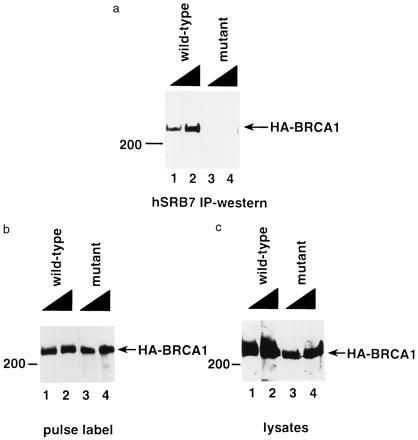
Deletion of 10 amino acid residues from the carboxyl terminus of BRCA1 results in reduced association with holoenzyme. The 293T cells were transfected with 10 μg (odd numbered lanes) or 20 μg (even numbered lanes) of plasmid expressing BRCA1 genes fused to the HA tag on the amino terminus. (a) Mutant BRCA1 (lanes 3 and 4) revealed a reduced association with the holoenzyme. Lysates prepared from transfected cells were immunoprecipitated with affinity-purified anti-hSRB7 antibody and subjected to SDS/PAGE, and the immunoblot was stained with anti-HA antibody. (b) Wild-type and mutant BRCA1 were equally well synthesized in transfected cells. Samples treated in parallel with the experiment in a were pulse-labeled with [35S]methionine-containing medium. Lysates were prepared and immunoprecipitated with the anti-HA antibody and subjected to SDS/PAGE. (c) Steady-state levels of transfected protein were determined by subjecting lysates of the transfected cells used in (a) to SDS/PAGE and then immunoblotting and probing for the HA tag.
DISCUSSION
Intact BRCA1 is a component of the RNA pol II holoenzyme in HeLa cell extracts. This has been shown by (i) copurification of a significant fraction (50%) of the BRCA1 in the cell with a complex containing pol II and the SRB10 homologue Cdk8; (ii) by coimmunopurification of BRCA1, hSRB7, pol II, TFIIF, TFIIE, and TFIIH; and (iii) by association of epitope-tagged wild-type BRCA1 with the hSRB7-containing complex in cells. The significance of these observations arises from the fact that full-length BRCA1 can be captured in close association with a set of vital components of the cellular transcription machinery. Thus with the observations that a carboxyl-terminal segment of BRCA1 has overt transcription activation function (6, 7), that a BRCA1 mutant lacking 11 carboxyl-terminal residues failed to associate with the holoenzyme, and that other holoenzyme components fused to dedicated DNA binding domains also act as transcriptional activators in vivo (10, 26), it seems fair to conclude that at least one innate function of BRCA1 is to affect the expression of one or more genes at the level of transcription. Indeed, although earlier results on the transactivation function of a severely truncated species of BRCA1 left open this possibility, the demonstration of the relatively tight association of intact BRCA1 and the holoenzyme lends credibility to the notion that BRCA1 normally affects transcription. How, if at all, this relates to its recently imputed role in the maintenance of genome integrity and the enaction of double-strand break repair and homologous recombination (8) is a mystery. Nevertheless, one can imagine a circumstance in which a protein that participates in one or more checkpoint control functions acts, at least in part, by controlling the expression of certain genes whose products do the work of checkpoint activation. p53 is such a protein, and like BRCA1, it too is the product of a tumor suppressor gene.
Many transcription factors, when fused to the GAL4 DNA binding domain, can activate transcription. However, this does not mean that they are all components of the holoenzyme. For example, two of the factors tested in Fig. 2, YY1 and RelA, regulate transcription by contacting p300 (27) and/or CREB binding protein (CBP; ref. 28), two key transcriptional coactivators. CBP is a component of the holoenzyme (29); YY1 and RelA are not, as shown herein. Thus, not all transcription activation proteins that contact the holoenzyme copurify with it. BRCA1 might, then, be viewed as a member of a different class of transcription activator proteins, because it is a stable component of the pol II holoenzyme.
For CBP, the activation of transcription by phospho-CREB appears to depend upon an interaction of CREB with CBP in the holoenzyme and also of a glutamine rich domain in CREB with TFIID (29). CBP, and its homologue, p300, serve to coactivate multiple transcription activators involved in regulated transcription reactions, including STAT2, nuclear hormone receptors, and NF-κB (28, 30–32), suggesting that these transcription activators contact p300/CBP, which, in turn, brings the holoenzyme into play. One wonders whether BRCA1 functions in an analogous or complementary manner, serving as a bridging protein between a regulator of transcription and the holoenzyme.
Acknowledgments
We thank E. Nigg, P. Rickert, E. Lees, Y. Shi, J. Aster, A. Neish, A. Roy, and D. Ginsberg for the kind gift of antibody reagents and M. Brown for suggesting BRCA1 may associate with the holoenzyme. This work was supported in part by National Institutes of Health Grant GM-53504, a research grant from the American Cancer Society, Massachusetts Division, and a Junior Faculty Research Award from the American Cancer Society to J.D.P.; a National Institutes of Health grant to D.M.L.; a Dana–Farber Women’s Cancer Program Fellowship to R.S.; and a institutional training grant from the National Institutes of Health to S.F.A.
ABBREVIATIONS
- pol II
RNA polymerase II
- SRB
suppressors of RNA polymerase B
- CTD
carboxyl-terminal domain
- HA
hemagglutinin
- CBP
CREB binding protein
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Futreal P A, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, et al. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 3.Szabo C I, King M C. Hum Mol Genet. 1995;4:1811–1817. doi: 10.1093/hmg/4.suppl_1.1811. [DOI] [PubMed] [Google Scholar]
- 4.Smith S A, Easton D F, Evans D G R, Ponder B A J. Nat Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 5.Neuhausen S L, Marshall C J. Cancer Res. 1994;54:6069. [PubMed] [Google Scholar]
- 6.Monteiro A N A, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 8.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 9.Koleske A M, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 10.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 11.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 12.Thompson C M, Young R A. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ossipow V, Tassan J P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 14.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 16.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 17.Tassan J P, Jaquenoud M, Leopold P, Schultz S J, Nigg E A. Proc Natl Acad Sci USA. 1995;92:8871–8875. doi: 10.1073/pnas.92.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E. Oncogene. 1996;12:2631–2640. [PubMed] [Google Scholar]
- 19.Manley J L, Fire A, Samuels M, Sharp P A. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 20.Seto E, Shi Y, Shenk T. Nature (London) 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 21.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Nature (London) 1991;354:254–258. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 22.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson N E, Aronson D B, Burgess R R. J Biol Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- 24.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 25.Scully R, Ganesan S, Brown M, DeCaprio J A, Cannistra S A, Feunteun J, Schnitt S, Livingston D M. Science. 1996;272:123–125. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- 26.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 27.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 28.Gerritsen M E, Williams A, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M R. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 31.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]


