Abstract
In yeast, the major mRNA degradation pathway is initiated by poly(A) tail shortening that triggers mRNA decapping. The mRNA is then degraded by 5′-to-3′ exonucleolysis. In mammalian cells, even though poly(A) tail shortening also precedes mRNA degradation, the degradation pathway has not been elucidated. We have used a reverse transcription–PCR approach that relies on mRNA circularization to measure the poly(A) tail length of four mammalian mRNAs. This approach allows for the simultaneous analysis of the 5′ and 3′ ends of the same mRNA molecule. For all four mRNAs analyzed, this strategy permitted us to demonstrate the existence of small amounts of decapped mRNA species which have a shorter poly(A) tail than their capped counterparts. Kinetic analysis of one of these mRNAs indicates that the decapped species with a short poly(A) tail are mRNA degradation products. Therefore, our results indicate that decapping is preceded by a shortening of the poly(A) tail in mammalian cells, as it is in yeast, suggesting that this mRNA degradation pathway is conserved throughout eukaryotic evolution.
Keywords: [mRNA stability, polyAtail, cap, reverse transcription–PCR]
Messenger RNA degradation contributes significantly to the regulation of gene expression. In eukaryotes, elucidation of a number of mRNA degradation pathways is under way (for reviews, see refs 1–4). Presently, these pathways are better understood in yeast than in mammalian cells. Both in yeast and in mammalian cells, degradation of most polyadenylylated mRNAs appears to be initiated by poly(A) shortening: transcriptional pulse–chase experiments have shown that shortening of the poly(A) tail precedes mRNA degradation (5, 6). Some regulatory sequence determinants affect mRNA stability by modulating the rate of deadenylylation, whereas others modulate a later degradation step (5–7).
This later step has been elucidated in yeast: deadenylylation at the 3′ end triggers mRNA decapping at the 5′ end, which is then followed by 5′-to-3′ exonucleolysis (refs. 8 and 9 and references therein). In some specific circumstances, other degradation pathways are observed: exonucleolysis from the 3′ end can occur when the 5′-to-3′ exonuclease is inactive, and a decapping pathway independent of poly(A) tail shortening is involved in the degradation of mRNAs that show premature translation termination (9–11).
In mammalian cells, the degradation pathway that follows deadenylylation is not well understood. Uncapped mRNAs are less stable than their capped counterparts in cell extracts, and enzymatic activities that catalyze mRNA decapping and 5′-to-3′ exonucleolysis have been identified (refs. 1 and 3 and references therein). Furthermore, there is a conservation between yeast and mammalian cells of a functional interaction between the 5′ and 3′ ends of mRNAs: in both systems, these two ends contribute to translational control (e.g., see refs. 12 and 13). It is thus tempting to speculate that deadenylylation triggers a decapping-dependent degradation pathway in mammalian cells as well. However, a direct demonstration is lacking.
We have developed a reverse transcription (RT)-PCR approach to measuring the length of the poly(A) tail, allowing us to examine the relationship between poly(A) tail length and the decapping event. This approach relies on RNA ligase-mediated mRNA circularization and thus allows for the simultaneous analysis of the 5′ and 3′ ends of the same mRNA molecule. Using this strategy to analyze four specific mammalian mRNAs, we detected minor amounts of decapped species and showed that they possess a much shorter poly(A) tail than the corresponding capped mRNAs. This indicates that decapping is triggered by a shortening of the poly(A) tail in mammalian cells.
MATERIALS AND METHODS
RNA Preparation.
Mouse liver RNA used for serum amyloid A (SAA) mRNA analysis was prepared from female CBA/J mice as follows: Acute inflammation was induced by subcutaneous injection of 0.1 ml of 10% azocasein in the lumbar region. At timed intervals (8 and 48 hr) after stimulus, animals were killed by cervical dislocation (the zero time point is an unstimulated animal). The liver was frozen in liquid nitrogen, and total cellular RNA was purified by the method of Auffray and Rougeon (14). Systemic inflammation was monitored and confirmed by determining serum amyloid P levels by rocket immunoelectrophoresis (15). Rat hepatoma cell (H4II or FTO2B) RNA was prepared using RNAXEL (Eurobio, Paris).
RNA samples were then treated with DNase I as follows: 50 μg of RNA was incubated with 20 units of human placental ribonuclease inhibitor (HPRI; Amersham) and 20 units of RNase-free DNase I (Pharmacia) in 100 μl of 50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2 for 20 min at 37°C. After extractions with phenol and chloroform, RNA was precipitated with ethanol.
Oligo(dT)/RNase H treatment was performed as follows: 10 μg of RNA was mixed with 500 ng of oligo(dT)12–18 (Pharmacia) in 20 μl of 200 mM KCl/1 mM EDTA, pH 8, incubated 2 min at 90°C, and annealed for 10 min at 25°C. The sample was then treated for 30 min at 37°C with 3 units of RNase H (Promega) and 20 units of HPRI in 40 μl of 20 mM Tris⋅HCl, pH 8/28 mM MgCl2/100 mM KCl/0.5 mM EDTA. After extractions with phenol and chloroform, RNA was precipitated with ethanol.
Circularization RT-PCR (cRT-PCR).
The procedure we used was derived from the procedures of Mandl et al. (16) and Bertrand et al. (17) and is depicted in Fig. 1.
Figure 1.
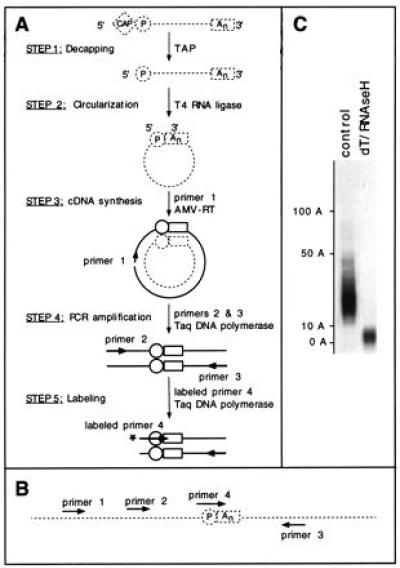
The cRT-PCR procedure. (A) Strategy used for measuring the poly(A) tail length of a specific mRNA. Messenger RNA molecules are represented as broken lines and DNA molecules as thick lines. The cap and poly(A) tail (An) are indicated by a diamond and a rectangle, respectively. The circle indicates the 5′-phosphate group of the most 5′ base of the original mRNA. AMV, avian myeloblastosis virus. (B) Arrangement of the primers used. The junction point of the circularized RNA is shown. (C) cRT-PCR allows one to measure the size distribution of the poly(A) tail. Albumin (ALB) mRNA poly(A) tail length was analyzed using hepatoma cells RNAs. The RNAs were decapped with tobacco acid pyrophosphatase (TAP) and subjected to cRT-PCR without (control lane), or with (dT/RNase H lane) removal of most of the poly(A) tail by treatment with oligo(dT) and RNase H. The length of the poly(A) tails as deduced by comparison to a size standard is indicated to the left. Labeling was performed during the PCR (25 cycles) using labeled primer 3.
Capped RNAs were decapped following a modification of the procedure of Fromont et al. (18): 10 μg of RNA was incubated with 2.5 units of tobacco acid pyrophosphatase (TAP; Epicentre Technologies, Madison WI) and 20 units of HPRI in 20 μl of 50 mM sodium acetate, pH 6/1 mM EDTA/1% 2-mercaptoethanol/0.1% Triton X-100 for 1 hr at 37°C and then precipitated with ethanol.
Circularization was performed as follows: 4 μg of RNA was incubated with 20 units of T4 RNA ligase (Pharmacia) and 20 units of HPRI in 400 μl of 50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/20 mM dithiothreitol/100 μM ATP/100 μg/ml acetylated BSA for 16 hr at 16°C. After extractions with phenol and chloroform, RNA was precipitated with ethanol.
Reverse transcription, PCR amplification, and labeling were performed as described for the reverse-ligation-mediated-PCR (RL-PCR) procedure (17) with the following modifications: MnCl2 was omitted from the PCR, PCR amplification was performed in 20 μl for 20 cycles [30 sec at 95°C, 3 min at the primer annealing temperature (see below), and 3 min at 74°C] except for the samples not treated with TAP, which were amplified for 25 cycles. For labeling of the products, 5 μl of the PCR mixture was further amplified for 5 cycles in a 10-μl reaction mixture containing 5 ng of the primer radiolabeled as described (17).
Primers.
The primers were prepared as described previously (17). The various primer combinations used are depicted in Fig. 1B. For all mRNAs analyzed, the reverse transcription reaction was performed using primer 1. To analyze SAA mRNA, we used primers 2 and 3 during the PCR amplification. For the results shown in Fig. 1C, primer 2 was radiolabeled to label the PCR products directly as described for the RL-PCR procedure (17). For the results shown in Fig. 2, we used radiolabeled primer 4 during the labeling step. To analyze tyrosine aminotransferase (TAT) mRNA, we used primers 1 and 3 during the PCR amplification and primer 4 during the labeling step. To analyze ALB and phosphoenolpyruvate carboxykinase (PCK) mRNAs, we used primers 1 and 3 during the PCR amplification and primer 4 during the labeling step.
Figure 2.
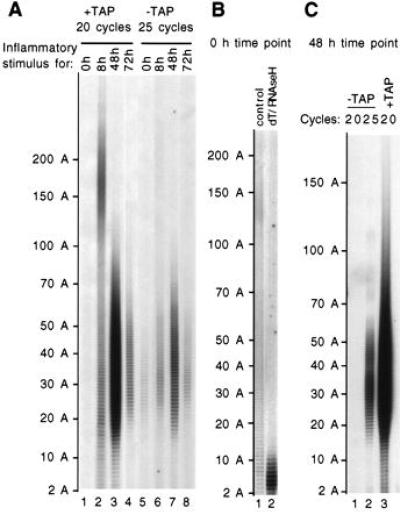
Evidence for traces of uncapped SAA mRNA with a shorter poly(A) tail than their capped counterparts. (A) Variation in SAA mRNA poly(A) tail length after induction of the acute-phase response was analyzed in liver RNAs of mice treated with azocasein for 0 (lanes 1 and 5), 8 (lanes 2 and 6), 48 (lanes 3 and 7), or 72 hr (lanes 4 and 8). Capped SAA mRNA species were analyzed after TAP treatment (+TAP, lanes 1–4), whereas the uncapped mRNA species were analyzed by omitting the TAP treatment (−TAP, lanes 5–8). In all cases, labeling of the PCR products was performed with labeled primer 4 for five additional cycles. The length of the poly(A) tails as deduced by comparison to a size standard is indicated to the left. (B) The size distribution of the poly(A) tail is heterogeneous without induction. A longer exposure of a gel performed using more material (four times) than in A is shown. SAA mRNAs from an untreated mouse were decapped with TAP and subjected to cRT-PCR without (control, lane 1), or with (dT/RNase H, lane 2) removal of most of the poly(A) tail by treatment with oligo(dT) and RNase H. (C) The material analyzed after TAP treatment corresponds only to the capped mRNA population. Using the mRNA from mice treated with azocasein for 48 hr, amplification was performed for 20 cycles for both the material treated or not with TAP (respectively, lanes 3 and 1) as well as for 25 cycles for the untreated material (lane 2).
The nucleotide sequences (5′ to 3′) of the primers were as follows (the annealing temperature used for each primer is indicated in parentheses): SAA primer 1, AGCAGGGAGCAGAAGA (42°C); SAA primer 2, GCTTCATCCTGCTGGTGTCTC (62°C); SAA primer 3, CTTGAGGAGGAGAGGGTAATA (62°C); SAA primer 4, GGCAGATCTGGTGGTCTATTT (62°C); TAT primer 1, GCAGCCTCCCAGCAGCC (56°C); TAT primer 3, TAGAATCATATATAGCCACTTTTG (56°C); TAT primer 4, CTCCACCAAGTCCTCTGATGATTT (68°C); ALB primer 1, AGGAGGAGGAGAAAGGTTAC (56°C); ALB primer 3, ACTCATCTCTTCTGTTGGTG (56°C); ALB primer 4, AGTGGGTTGACAGGAAAGGTGGTTT (70°C); PCK primer 1, TCTCAGAGCGTCTCGCCG (56°C); PCK primer 3, ATAATTGAAATAGGTATCCTGAC (56°C); and PCK primer 4, CCAGAGGGAAGGCCAACCGTTT (70°C).
Controls.
To assess the specificity of the signal observed, we perform the following controls. (i) We verify that the signal is not due to DNA contamination by omitting the reverse transcriptase in the primer extension step. (ii) We verify that the ligation has occurred in vitro by omitting the RNA ligase in the ligation step. (iii) When possible, the authenticity of the signal is verified by restriction enzyme digestion of the PCR product (performed here for the TAT and PCK mRNA). (iv) Even though it is unlikely that the signal originates from intermolecular ligation of two mRNA molecules originating from the same gene, given their usually low abundance, we verified that, in the ligation conditions used, such an event occurs at a frequency too low to contribute to the signal by using a primer 3 and a primer 4 from different mRNA species. In addition, it is unlikely that the signals analyzed here could originate from a 5′ poly(A) tract, as observed for vaccinia virus mRNAs (e.g., see ref. 19), because the 5′ end of three of the mRNA species analyzed here (namely SAA2, TAT, and PCK) has been determined by both S1 nuclease mapping and primer extension (20–22).
RESULTS
A PCR Approach to Measure the Poly(A) Tail Length of a Specific mRNA.
We have used an experimental strategy derived from the procedure described by Mandl et al. (16). The basic principle is the circularization of RNA molecules by using RNA ligase, cDNA synthesis across the ligated termini, and PCR amplification of the poly(A) tail and surrounding sequences (Fig. 1). The cRT-PCR requires that the cap structure at the 5′ end be absent or removed. In the first step, cap removal is catalyzed by TAP (16, 18). In the second step, the 5′ and 3′ ends of the RNA are intramolecularly ligated, using T4 RNA ligase at a low RNA concentration. When performed using total cellular RNA, these first two steps should occur unspecifically on every RNA species. Specificity is provided in the subsequent steps using a set of nested primers as described for the RL-PCR procedure (17). A cDNA copy of the circularized RNA of interest is made, using primer 1 and avian myeloblastosis virus reverse transcriptase (step 3, Fig. 1A). This allows for the synthesis of a copy of the poly(A) tail that is ligated to the 5′ end. This poly(A) tail and surrounding sequences are amplified by using Taq DNA polymerase and two primers that hybridize on both sides of the junction region (step 4, Fig. 1A). The pool of amplified PCR products is visualized on a denaturing polyacrylamide gel after extension of a 5′-end-labeled nested primer (step 5, Fig. 1A). The size distribution of the poly(A) tail is deduced by comparison to a size standard and subtraction of the length of the sequence surrounding the poly(A) tail.
Fig. 1C shows the results of a cRT-PCR analysis of the poly(A) tail of the mRNA coding for rat ALB. This analysis shows the resolution power of the cRT-PCR to measure the size distribution of the poly(A) tail and reveals that most of the mRNA in this sample has a poly(A) tail that ranges from 10 to 50 adenosines. After a combined oligo(dT)/RNase H treatment, the length of this tail is reduced to between 0 and 11 adenosines, showing that the size distribution of the PCR products does reveal the size distribution of the poly(A) tail. The incomplete elimination of the poly(A) tract presumably results from a combination of the length of the oligo(dT) (12–18 nucleotides) and the temperature of RNase H treatment (37°C): mRNAs that have a poly(A) tail shorter than 12 As might not allow further hybridization of oligo(dT).
Decapped SAA mRNA Species Have a Short Poly(A) Tail.
The products of a subset of SAA-superfamily genes (SAA1, -2, and -3) are major acute-phase reactants and apolipoproteins of high density lipoprotein (for a review, see ref. 23). In liver, the transcription of these SAA genes is strongly stimulated in response to inflammatory stimuli. The mRNA of both SAA genes accumulates during the first 24 hr of stimulation, then return to basal level about 72 hr after the stimulating event (15, 24). Northern blot analyses have shown that the mRNA poly(A) tail length varies during this period from 150 to 200 bases early after stimulation (6–8 hr) to 50 to 100 bases 36 hr after stimulation (15, 24). These observations are compatible with a limited transcriptional pulse of SAA genes and a regular poly(A) tail shortening preceding mRNA degradation, as described for other mRNAs analyzed in transcriptional pulse–chase experiments (5, 6).
We have used the cRT-PCR procedure to analyze the variations in poly(A) tail length of SAA2 mRNA in the liver of mice during the acute-phase response (Fig. 2A). Eight hours after stimulation, most of the SAA2 mRNA poly(A) tail ranges in size from 100 to 300 As, with a peak around 170 As (lane 2), whereas 48 hr after stimulation, this size range has decreased to 20 to 70 As (lane 3). Seventy-two hours after stimulation the signal has markedly decreased due to mRNA degradation (lane 4). Before stimulation, the poly(A) tail of the traces of mRNAs that can be detected has a more heterogeneous size distribution, ranging from 20 to 300 As (visible on a longer exposure of the gel; Fig. 2B, lane 1). The specificity of this faint heterogeneous signal is assessed by an oligo(dT)/RNase H treatment which brings down all material to a poly(A) length shorter than 11 As (Fig. 2B, lane 2). The heterogeneous size distribution of the poly(A) tail in the absence of stimulation is presumably due to the age heterogeneity of the mRNA resulting from continuous basal transcription of the gene.
While using the RL-PCR procedure to map the 5′ ends of various transcripts (18), we have regularly observed the presence of traces of full-length decapped transcripts onto which the 3′-OH end of an RNA linker could be ligated without a prior TAP treatment (unpublished data). Such full-length 5′-P mRNA species could be degradation products of the mRNA. Since the cRT-PCR procedure allows us to simultaneously analyze the 5′ and 3′ ends of the same mRNA molecule, we have measured the length of the poly(A) tail of these full-length decapped species simply by omitting the first step of the procedure: the TAP treatment (see Fig. 1). Five more cycles were performed during the PCR amplification to detect these less abundant decapped species. To distinguish the decapped mRNAs from those resulting from more extensive degradation (e.g., either exonucleolytic or endonucleolytic cleavages), we visualized the PCR products by using a labeled primer (no. 4) that is complementary to the junction point—i.e., complementary to the 5′ end of the mRNA and containing in addition two thymidine residues at its 3′ end capable of hybridizing with the last two adenosine residues of the poly(A) tail (see Fig. 1B). This ensures that the PCR products detected correspond only to the oligo(A) species that have a complete decapped 5′ end (the first nucleotide could have been lost and this loss compensated by the 3′ end, because this first nucleotide is an adenylate residue).
Using this strategy, we observe that the traces of decapped SAA mRNA that can be detected have a short poly(A) tail (from 20 to 60 As) throughout the acute-phase response (lanes 5–8; Fig. 2A). This is true even at 8 hr after stimulation, when the bulk of the capped mRNA population has a mean poly(A) tail length of 170 As (compare lanes 2 and 6). The relative amount of decapped species throughout the acute-phase response indicates that they are degradation products of the capped mRNAs. The level detected is similar at early time points (0 and 8 hr) when the degradation of the bulk of the induced mRNA has not yet been initiated; it rises at 48 hr in parallel with the decrease of the poly(A) length; and it decreases at 72 hr when most of the induced mRNA has been degraded. The signal analyzed here with the TAP-treated material originates essentially only from the capped mRNA population. Indeed, when the material untreated with TAP is analyzed using the same number of amplification cycles (20), it does not give rise to a significant signal (compare lanes 1 and 3 in Fig. 2C). At 8 hr after stimulation, a significant proportion of the capped mRNA population has a short poly(A) tail (shorter than 70 As, lane 2, Fig. 2A). This suggests that a part of the SAA mRNA population could be deadenylylated faster than the bulk.
In conclusion, the detection of decapped species with a short poly(A) tail indicates that SAA mRNA decapping follows deadenylylation as described for some yeast mRNAs (6).
Traces of Decapped Species with a Short Poly(A) Tail Can Be Detected for Three Other Liver mRNAs.
To determine whether these observations extend to other mammalian mRNAs, we used the cRT-PCR procedure to compare the steady-state poly(A) tail length of capped and decapped species of three other liver mRNAs, those coding for PCK, ALB, and TAT (Fig. 3). Analysis of PCK mRNA poly(A) tail length shows a tract ranging from 5 to 100 nucleotides, with a peak around 25 adenylate residues, whereas that of the corresponding decapped species varies from 2 to 21 adenylate residues (with a peak around 7). The poly(A) tail length of ALB mRNA ranges from 10 to 50 nucleotides (with a peak around 25), whereas that of the decapped species varies from 4 to 24 adenylate residues (with a peak around 15). For TAT mRNA, the poly(A) tract is short, ranging from 2 to 50 nucleotides with a peak at 10 nucleotides, while that of the decapped counterpart ranges from 2 to 12 adenylate residues (with a peak around 7). These results show that, for all mRNAs analyzed, the poly(A) tail of the decapped species is shorter than that of their capped counterparts. These results provide further evidence that deadenylylation precedes decapping and that deadenylylation-dependent decapping could be a general degradation pathway in mammalian cells, as it is in yeast cells.
Figure 3.
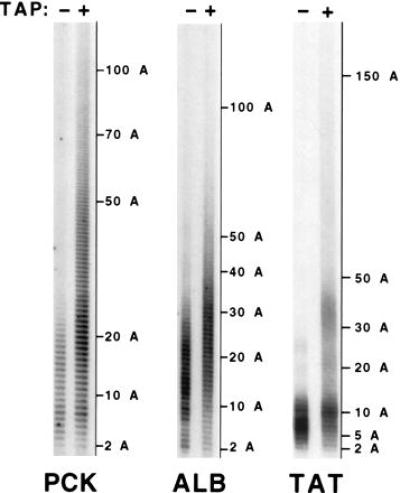
Traces of uncapped mRNA species with short poly(A) tails can also be detected for three other mRNAs. PCK, ALB, and TAT mRNAs were analyzed as described for SAA mRNA in Fig. 2, except that (i) the RNA preparation originated from unstimulated rat hepatoma cells, (ii) the amplifications were performed with primers 1 (i.e., the reverse transcription primer) and 3, and (iii) the number of amplification cycles performed to analyze ALB mRNA was 25 and 29, respectively, for the capped and uncapped species.
DISCUSSION
cRT-PCR: A Sensitive Method to Measure Poly(A) Tail Size Distribution.
We have developed an RT-PCR-based assay to measure the size distribution of the poly(A) tail of mRNAs. The use of PCR renders this method much more sensitive than methods using RNase H treatment and Northern blotting (e.g., see ref. 25). The use of nested primers and the visualization of the PCR products with a labeled primer, as described for the LM-PCR and RL-PCR procedures (17, 26), allows direct analysis of the whole poly(A) tail population, in contrast to alternative procedures for RNA ligase-mediated amplification of cDNA ends (16, 27), which relies on DNA cloning and sequencing of the PCR products. In cRT-PCR, the mRNA is circularized prior to reverse transcription of the poly(A) tail, using a primer that hybridizes to the 5′ end of the original transcript. Other RT-PCR-derived procedures rely on the use of a tagged oligo(dT) primer (e.g., ref. 28). Since, in such cases, the primer can hybridize internally to the poly(A) tail, these procedures are useful for determining maximum poly(A) tail length in an mRNA population but the PCR products do not accurately reflect the size distribution of the population. A recently described modification of this procedure circumvents this problem (29). It relies on the ligation of oligo(dT) and an oligo(dT)-tagged primer annealed to the poly(A) tail followed by RT-PCR. The specific advantage to the cRT-PCR approach described herein is that it allows for the simultaneous analysis of the 5′ and 3′ ends of the same mRNA molecule. This permitted us to detect, in mammalian cells, decapped mRNAs with short poly(A) tails.
Deadenylylation-Dependent Decapping of SAA mRNA.
We have used cRT-PCR to analyze the variation in size distribution of the poly(A) tail of SAA2 mRNA during the acute-phase response in mouse liver. The transcription of these genes is induced by the concerted action of glucocorticoids, interleukin 1, interleukin 6, tumor necrosis factor α, and other hormones during inflammation (23). The poly(A) tail length of the mRNA varies as if there was a limited pulse of transcription induced by these hormones. Eight hours after induction, newly synthesized mRNAs have a long poly(A) tail (100–300 residues), whereas 48 hr later, the average length has shortened to 50 residues. This shortening precedes the disappearance of the mRNA (24). During the inflammatory response, decapped SAA2 mRNAs with short poly(A) tails (20–60 residues) were detected. The relative proportion of decapped versus capped mRNA species was not determined accurately. At least five extra amplification cycles are necessary to detect similar amounts of PCR products from the decapped species. This suggests that the amount of decapped species represents 3% of that of their capped counterparts, assuming a 100% efficiency of conversion of capped mRNA to the 5′-monophosphorylated form during the TAP reaction. However, in our hands, the in vitro decapping efficiency is somewhat variable, and this 3% proportion is probably an overestimate. During the inflammatory response, the decapped SAA2 mRNA appears more abundant at later times, when the bulk of the mRNA synthesized in response to the inflammatory stimuli begins to be degraded. This suggests that the decapped species is an intermediate degradation product of the mRNA. In yeast cells, mRNA degradation can proceed through deadenylylation-triggered decapping, followed by 5′-to-3′ exonucleolysis (6). The enzymes responsible for decapping and 5′-to-3′ exonucleolysis copurify, and it is tempting to speculate that they interact in vivo to allow coordination of these successive degradation steps (30). Indeed, nearly full-length decapped species have not been observed without the use of genetic tricks to stabilize these intermediates: either insertion of an exonuclease-resistant structure in the 5′ end or genetic inactivation of the major 5′-to-3′ exonuclease gene (6). Thanks to the sensitivity of the cRT-PCR procedure, we have been able to directly detect such intermediates in mammalian cells. Late after induction, the decapped SAA mRNA species is slightly more abundant, but it does not accumulate extensively, presumably because of the instability of this degradation intermediate.
It is unlikely that the decapped species with a short poly(A) tail that we detect are produced during synthesis of SAA mRNA and not during its degradation. Indeed, this would have resulted in an inverse accumulation pattern of this species, with high level detected early during induction. Furthermore, current knowledge of the polyadenylylation process does not favor this possibility. Polyadenylylation in mammalian nuclei occurs in two phases: an initial extension of a 10- to 20-A tail stimulated by the AAUAAA recognition complex, followed by a rapid polymerization of up to 200 adenylates, which is stimulated by the nuclear polyadenylate-binding protein (PABP2) that binds to the previously synthesized oligo(A) tail (see refs. 31 and 32 for reviews). Decapped species with an oligo(A) tail could have been generated during synthesis if the second poly(A) addition phase were dependent upon the presence of the cap, in contrast to the previous mRNA biosynthetic and maturation steps. This is not what is observed in in vitro polyadenylylation reactions: the cap does not influence the rate of the first phase of polyadenylylation and is not required for the second (33). Furthermore, the cap plays an important role in the splicing of the first intron (34). Since one of the primers we have used (no. 2) overlaps the exon 1–exon 2 boundary, we analyzed only transcripts where the first intron has been spliced and thus, most likely, transcripts that have been capped. Therefore, we conclude that the oligoadenylated decapped mRNA species that we observe are degradation products of SAA mRNA.
Generality of the Deadenylylation-Dependent Decapping in Mammals.
We have analyzed the poly(A) size distribution of four liver mRNAs of various stabilities. While both SAA and ALB mRNA could be considered as stable [half-life > 24 hr (24, 35)], TAT and PCK are unstable [half-life between 30 min and 2 hr (36, 37)]. Decapped species were detected for all these mRNAs and they always have a shorter poly(A) tail than their capped counterparts, suggesting the generality of the deadenylylation-dependent decapping pathway irrespective of the stability of the mRNA [as observed in yeast (6)]. However, the size distribution of the poly(A) tail of these various mRNAs differs at steady state. For the capped species, the average poly(A) tail lengths rank in the following order: SAA mRNA, 20–300 As (in the absence of induction); PCK mRNA, 5–100 As; ALB mRNA, 10–50 As; TAT mRNA, 2–50 As. While it cannot be formally excluded that these differences could result from differences in the length of the poly(A) tail added in the nucleus (see ref. 31 for a review), it is likely that they result from differences in the relative rates of deadenylylation and the subsequent degradation steps. Both in yeast and in mammalian cells, instability determinants, located either in the 3′ untranslated region or in the coding region of the mRNA, have been shown to independently affect these degradation steps (5–7). If deadenylylation proceeds faster than the subsequent degradation, the average steady-state poly(A) tail length should be shorter than in the reverse situation. Furthermore, the threshold minimal poly(A) tail length at which the subsequent decapping occurs might differ for individual mRNAs. This could account for the differences in the poly(A) size distribution of the in vivo decapped forms of the various mRNAs analyzed, which ranges from 20 to 60 As for SAA mRNA, from 2 to 21 As for PCK mRNA, from 4 to 24 As for ALB mRNA, and from 2 to 12 As for TAT mRNA. Except, to a small extent, for ALB mRNA, the minimal length of the poly(A) tail of the decapped species is not shorter than that of the capped species. This suggests that the final deadenylylation occurs at a slower rate than the deadenylylation-triggered event that affects the 5′ end. Indeed, our primer selection strategy precludes amplification of the mRNA molecules that have lost any nucleotides at the 5′ end as well as mRNAs with a 3′ end containing less than 2 to 3 As (for SAA2, TAT, and PCK mRNAs, the 5′-end base could have been lost and this loss compensated by the 3′ end, because this 5′-end is an adenylate residue). For SAA mRNA, it is clear that the final deadenylylation event occurs at a much slower rate than the deadenylylation-triggered decapping and degradation, since no significant amount of mRNA with a poly(A) tail shorter than 20 As is detected, even in the decapped species. Shortening of the poly(A) tail down to about 60 As appears to be able to trigger decapping, since it is the maximal length of the poly(A) tail of the in vivo decapped species. Assuming, as discussed previously, that deadenylylation of SAA mRNA does not proceed significantly further once decapping is triggered, the heterogeneity in poly(A) tail length of the decapped species may reflect the heterogeneity of the threshold length that triggers decapping of this mRNA. ALB mRNA shows decapped species with an oligo(A) tail as long as 24 As, indicating that it has a lower threshold length that triggers decapping. Furthermore, the minimal length of this oligo(A) tail is shorter (4 As), and shorter than that of the corresponding capped mRNA, suggesting that terminal deadenylylation is still proceeding before completion of 5′-end decapping and exonucleolysis. For PCK and TAT mRNAs, fully deadenylylated capped species may exist, even though the primer design strategy excludes them, because the poly(A) tail size distribution is homogeneous down to the lower limit of the analysis. Capped TAT mRNA species show a strikingly short poly(A) tail: the average size is 7 As and mRNAs with 3 As can be detected. Furthermore, decapped mRNAs do not have a poly(A) tail longer than 12 residues. These observations suggest that deadenylylation proceeds relatively faster than decapping when compared with the other mRNAs analyzed herein, and/or that the threshold oligo(A) length that triggers decapping is short. In conclusion, the varying size distribution of the capped and decapped species of the mRNAs analyzed here reveals the diversity of the kinetic parameters of the various steps of the common degradation pathway that affects these mRNAs.
The cytoplasmic PABP (PABP1) is likely to be one of the factors that affect these various deadenylylation and decapping processes. It is highly conserved from yeast to mammals (38), and it appears to protect mRNA from degradation both in yeast (39) and in mammalian cell extracts (40). In yeast, PABP1 appears to participate in the regulation of the rate of deadenylylation and to protect the mRNA from decapping (39). Deadenylylation could trigger decapping by eliminating PABP1 binding sites on the mRNA. Moreover, PABP1 serves as a cofactor for one of the enzymes involved in deadenylylation, the poly(A) ribonuclease (PAN) product of the Pan2 gene (41). In vitro, using purified PAN and PABP1, it has been shown that PAN-dependent deadenylylation can be regulated by sequences present in the 3′ untranslated region (UTR) of mRNAs (42). Since PABP1 is also able to interact with some 3′-UTR sequences (43), it could participate in the control of the rate of deadenylylation and eventually deadenylylation-dependent decapping, as well as in determining the threshold length at which deadenylylation triggers decapping (39, 40, 42). While considering the hypothesis that PABP1 is involved in the control of the threshold poly(A) length that triggers decapping in mammalian cells, it should be noted that the upper limit of this threshold length for SAA mRNA (60 residues) should be sufficient to allow for binding of two PABP1 molecules (44). This suggests that decapping is not an all-or-nothing decision, but rather is the consequence of an equilibrium between two antagonistic activities: that of the decapping enzyme and of its inhibitor, PABP1. The striking conservation of both PABP1 (38) and the deadenylylation-dependent decapping pathway from yeast to mammals suggests that PABP1 plays a central role in the control of mRNA stability in mammalian cells as in yeast.
Acknowledgments
We thank C. Dubucs for oligonucleotide synthesis. This work was supported in part by the Centre National de la Recherche Scientifique (R.P. and T.G.) and grants from the Association de Recherche sur le Cancer (R.P. and T.G.), the Ligue Nationale and the Ligue Parisienne contre le Cancer (R.P. and T.G.), a project grant from the Health Research Board of Ireland (D.S.) and a grant (no. 3160) from the Council for Tobacco Research—USA Inc. (D.S.). D.S. was supported by a Postdoctoral Research Fellowship from the Wellcome Trust, and P.C. was supported by a Fellowship from the Ministère de la Recherche et de l’Enseignement Supérieur.
ABBREVIATIONS
- RT-PCR
reverse transcription–PCR
- cRT-PCR
circularization–RT-PCR
- RL-PCR
reverse-ligation-mediated PCR
- SAA
serum amyloid A
- HPRI
human placental ribonuclease inhibitor
- TAP
tobacco acid pyrophosphatase
- ALB
albumin, PCK, phosphoenolpyruvate carboxykinase
- TAT
tyrosine aminotransferase
- PABP
polyadenylate-binding protein
- PAN
poly(A) ribonuclease
References
- 1.Beelman C A, Parker R. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 2.Peltz S W, Jacobson A. In: Control of Messenger RNA Stability. Belasco J G, Brawerman G, editors. San Diego: Academic; 1993. pp. 291–328. [Google Scholar]
- 3.Ross J. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sachs A B. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 5.Shyu A B, Belasco J G, Greenberg M E. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 6.Decker C J, Parker R. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 7.Chen C Y, Chen T M, Shyu A B. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. Nature (London) 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 9.Muhlrad D, Decker C J, Parker R. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 10.Muhlrad D, Decker C J, Parker R. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muhlrad D, Parker R. Nature (London) 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 12.Tarun S J, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 13.Gallie D R. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 14.Auffray C, Rougeon F. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 15.Zahedi K, Whitehead A S. J Immunol. 1989;143:2880–2886. [PubMed] [Google Scholar]
- 16.Mandl C W, Heinz F X, Puchhammer-Stockl E, Kunz C. BioTechniques. 1991;10:484–486. [PubMed] [Google Scholar]
- 17.Bertrand E, Fromont-Racine M, Pictet R, Grange T. Proc Natl Acad Sci USA. 1993;90:3496–3500. doi: 10.1073/pnas.90.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromont-Racine M, Bertrand E, Pictet R, Grange T. Nucleic Acids Res. 1993;21:1683–1684. doi: 10.1093/nar/21.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, Visca P, Vos J C, Stunnenberg H G. Cell. 1987;50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto K I, Goto N, Kosaka J, Shiroo M, Yeul Y D, Migita S. J Immunol. 1987;139:1683–1688. [PubMed] [Google Scholar]
- 21.Grange T, Guenet C, Dietrich J B, Chasserot S, Fromont M, Befort N, Jami J, Beck G, Pictet R. J Mol Biol. 1985;184:347–50. doi: 10.1016/0022-2836(85)90386-9. [DOI] [PubMed] [Google Scholar]
- 22.Beale E G, Chrapkiewicz N B, Scoble H A, Metz R J, Quick D P, Noble R L, Donelson J E, Biemann K, Granner D K. J Biol Chem. 1985;260:10748–10760. [PubMed] [Google Scholar]
- 23.Steel D M, Whitehead A S. Immunol Today. 1994;15:81–88. doi: 10.1016/0167-5699(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 24.Steel D M, Rogers J T, DeBeer M C, DeBeer F C, Whitehead A S. Biochem J. 1993;291:701–707. doi: 10.1042/bj2910701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vournakis J N, Efstratiadis A, Kafatos F C. Proc Natl Acad Sci USA. 1975;72:2959–2963. doi: 10.1073/pnas.72.8.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller P R, Wold B. Science. 1989;246:780–785. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Gorovsky M A. Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sallés F J, Darrow A L, O’Connel M L, Strickland S. Genes Dev. 1992;6:1202–1212. doi: 10.1101/gad.6.7.1202. [DOI] [PubMed] [Google Scholar]
- 29.Sallés F J, Strickland S. PCR Methods Appl. 1995;4:317–321. doi: 10.1101/gr.4.6.317. [DOI] [PubMed] [Google Scholar]
- 30.Stevens A. In: Control of Messenger RNA Stability. Belasco J G, Brawerman G, editors. San Diego: Academic; 1993. pp. 449–471. [Google Scholar]
- 31.Baker E J. In: Control of Messenger RNA Stability. Belasco J G, Brawerman G, editors. San Diego: Academic; 1993. pp. 367–415. [Google Scholar]
- 32.Wahle E, Keller W. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 33.Bienroth S, Keller W, Wahle E. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 35.Johnson T R, Rudin S D, Blossey B K, Ilan J, Ilan J. Proc Natl Acad Sci USA. 1991;88:5287–5291. doi: 10.1073/pnas.88.12.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernest M J. Biochemistry. 1982;21:6761–6767. doi: 10.1021/bi00269a022. [DOI] [PubMed] [Google Scholar]
- 37.Hod Y, Hanson R W. J Biol Chem. 1988;263:7747–7752. [PubMed] [Google Scholar]
- 38.Grange T, Martins de Sa C, Oddos J, Pictet R. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caponigro G, Parker R. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein P, Peltz S W, Ross J. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeck R, Tarun S J, Rieger M, Deardorff J A, Muller A S, Sachs A B. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 42.Lowell J E, Rudner D Z, Sachs A B. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- 43.Goldenberg S, Vincent A, Scherrer K. Nucleic Acids Res. 1980;8:5057–5070. doi: 10.1093/nar/8.21.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baer B W, Kornberg R G. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]


