Abstract
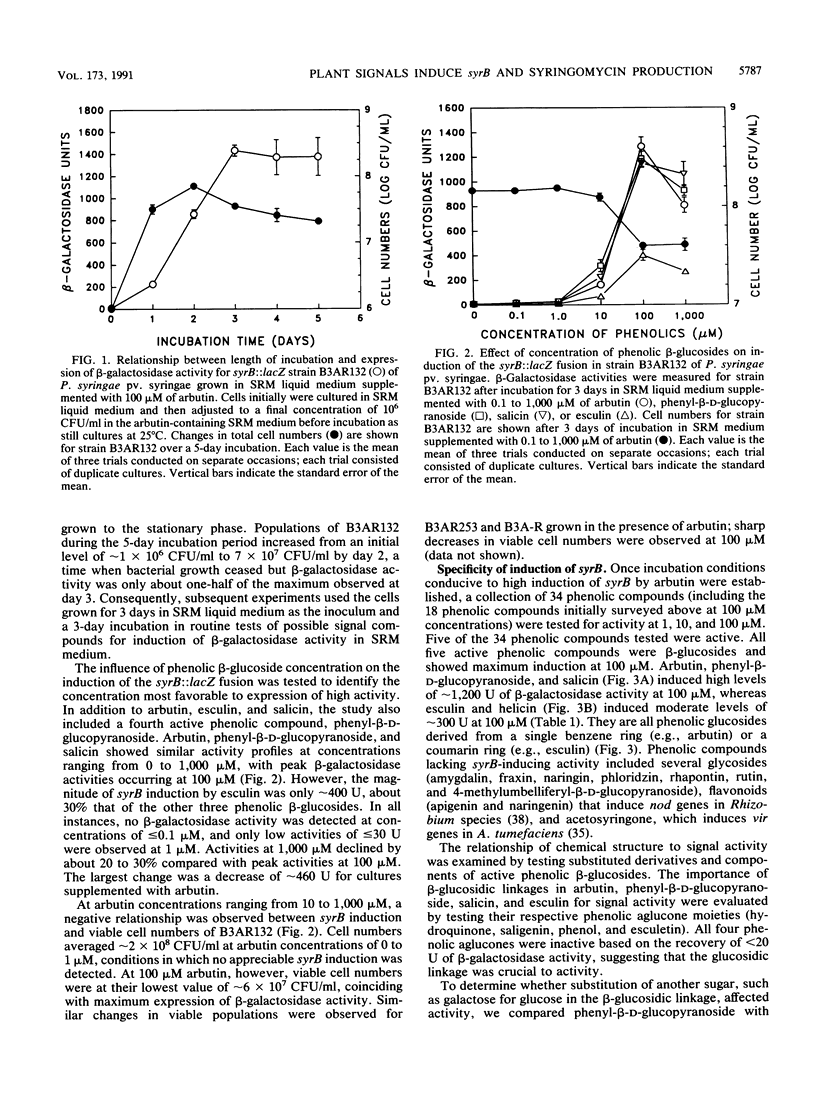
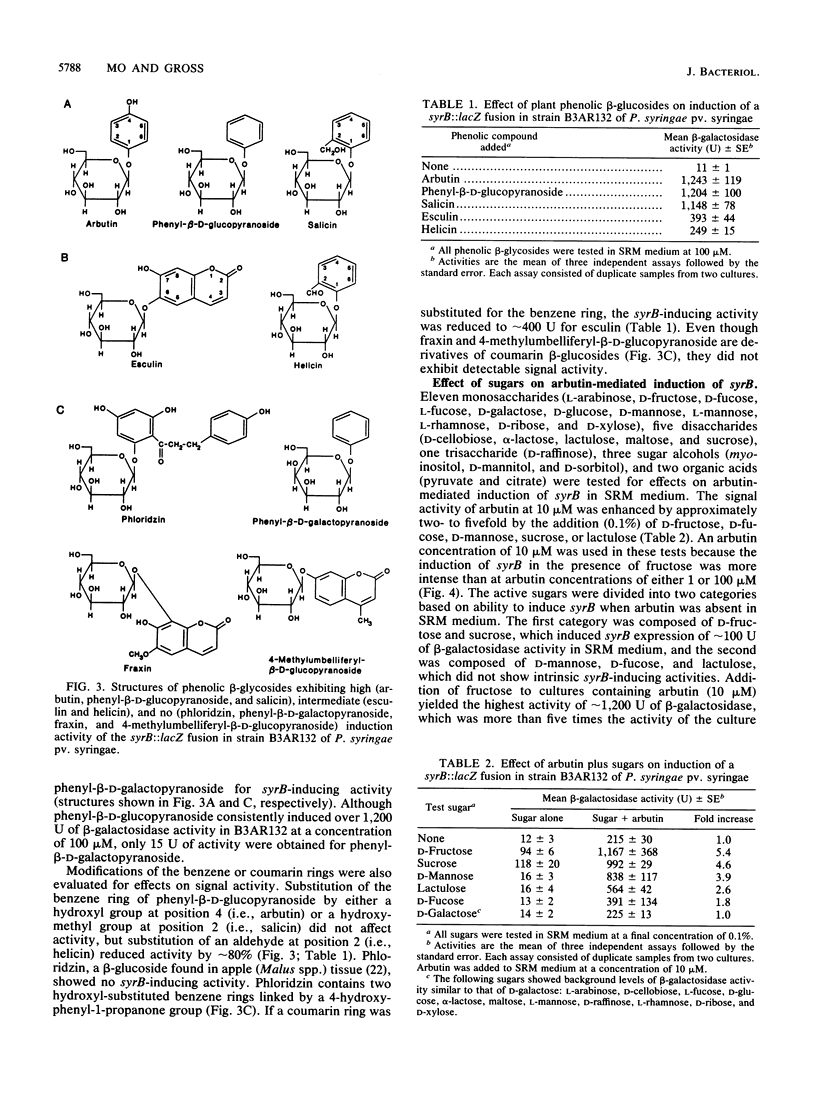
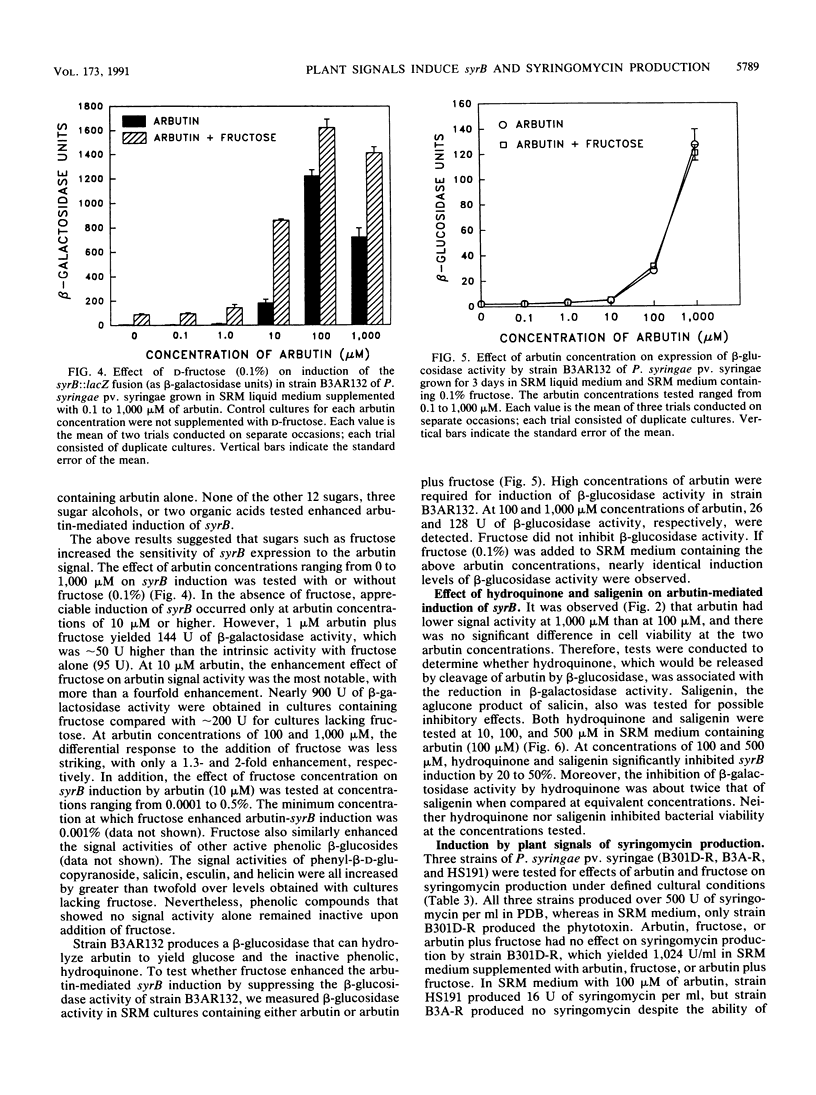
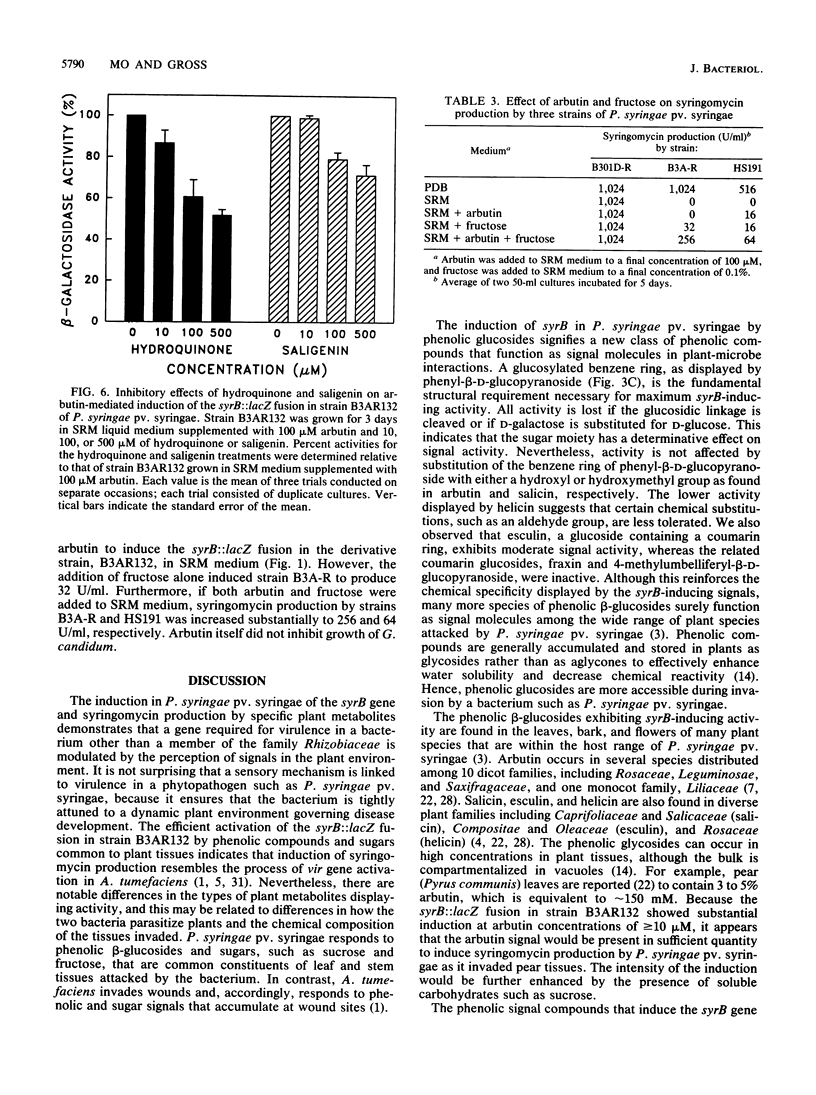
The syrB gene is required for syringomycin production by Pseudomonas syringae pv. syringae and full virulence during plant pathogenesis. Strain B3AR132 containing a syrB::lacZ fusion was used to detect transcriptional activation of the syrB gene in syringomycin minimal medium by plant metabolites with signal activity. Among 34 plant phenolic compounds tested, arbutin, phenyl-beta-D-glucopyranoside, and salicin were shown to be strong inducers of syrB, giving rise to approximately 1,200 U of beta-galactosidase activity at 100 microM; esculin and helicin were moderate inducers, with about 250 to 400 U of beta-galactosidase activity at 100 microM. Acetosyringone and flavonoids that serve as signal molecules in Agrobacterium and Rhizobium species, respectively, did not induce the syrB::lacZ fusion. All syrB inducers were phenolic glucosides and none of the aglucone derivatives were active, suggesting that the beta-glycosidic linkage was necessary for signal activity. Phenyl-beta-D-galactopyranoside containing galactose substituted for glucose in the beta-glycosidic linkage also lacked inducer activity. Phenolic signal activity was enhanced two- to fivefold by specific sugars common to plant tissues, including D-fructose, D-mannose, and sucrose. The effect of sugars on syrB induction was most noticeable at low concentrations of phenolic glucoside (i.e., 1 to 10 microM), indicating that sugars such as D-fructose increase the sensitivity of P. syringae pv. syringae to the phenolic plant signal. Besides induction of syrB, syringomycin biosynthesis by parental strain B3A-R was induced to yield over 250 U of toxin by the additions of arbutin and D-fructose to syringomycin minimal medium. These data indicate that syringomycin production by most strains of P. syringae pv. syringae is modulated by the perception of two classes of plant signal molecules and transduced to the transcriptional apparatus of syringomycin (syr) genes such as syrB.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer R. G., Nester E. W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990 Nov;172(11):6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwai A. P., Zhang L., Bachmann R. C., Takemoto J. Y. Mechanism of Action of Pseudomonas syringae Phytotoxin, Syringomycin : Stimulation of Red Beet Plasma Membrane ATPase Activity. Plant Physiol. 1987 Jan;83(1):39–43. doi: 10.1104/pp.83.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangelosi G. A., Ankenbauer R. G., Nester E. W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M. A., Redmond J. W., Batley M., Rolfe B. G. Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO J. 1987 May;6(5):1173–1179. doi: 10.1002/j.1460-2075.1987.tb02351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. C. Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J Appl Bacteriol. 1985 Feb;58(2):167–174. doi: 10.1111/j.1365-2672.1985.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert J. J., Hildebrand D. C., Schroth M. N. Nonutilization of beta-glucosides for growth by fluorescent pseudomonads. Phytopathology. 1970 Mar;60(3):502–505. doi: 10.1094/phyto-60-502. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kleinkauf H., von Döhren H. Biosynthesis of peptide antibiotics. Annu Rev Microbiol. 1987;41:259–289. doi: 10.1146/annurev.mi.41.100187.001355. [DOI] [PubMed] [Google Scholar]
- Melchers L. S., Regensburg-Tuïnk A. J., Schilperoort R. A., Hooykaas P. J. Specificity of signal molecules in the activation of Agrobacterium virulence gene expression. Mol Microbiol. 1989 Jul;3(7):969–977. doi: 10.1111/j.1365-2958.1989.tb00246.x. [DOI] [PubMed] [Google Scholar]
- Morgan M. K., Chatterjee A. K. Genetic organization and regulation of proteins associated with production of syringotoxin by Pseudomonas syringae pv. syringae. J Bacteriol. 1988 Dec;170(12):5689–5697. doi: 10.1128/jb.170.12.5689-5697.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. W., Morris R. O. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc Natl Acad Sci U S A. 1990 May;87(9):3614–3618. doi: 10.1073/pnas.87.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott K. A., Takemoto J. Y. Syringomycin, a bacterial phytotoxin, closes stomata. Plant Physiol. 1989 Aug;90(4):1435–1439. doi: 10.1104/pp.90.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N. K., Verma D. P. Phenolic compounds as regulators of gene expression in plant-microbe relations. Mol Plant Microbe Interact. 1990 Jan-Feb;3(1):4–8. doi: 10.1094/mpmi-3-004. [DOI] [PubMed] [Google Scholar]
- Segre A., Bachmann R. C., Ballio A., Bossa F., Grgurina I., Iacobellis N. S., Marino G., Pucci P., Simmaco M., Takemoto J. Y. The structure of syringomycins A1, E and G. FEBS Lett. 1989 Sep 11;255(1):27–31. doi: 10.1016/0014-5793(89)81054-3. [DOI] [PubMed] [Google Scholar]
- Shimoda N., Toyoda-Yamamoto A., Nagamine J., Usami S., Katayama M., Sakagami Y., Machida Y. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogn J. A. Structure of the peptide antibiotic polypeptin. J Med Chem. 1976 Oct;19(10):1228–1231. doi: 10.1021/jm00232a012. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., An G., Flores C., Nester E. W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985 Apr;4(4):891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. W., Gross D. C. Evaluation of the Role of Syringomycin in Plant Pathogenesis by Using Tn5 Mutants of Pseudomonas syringae pv. syringae Defective in Syringomycin Production. Appl Environ Microbiol. 1988 Jun;54(6):1345–1353. doi: 10.1128/aem.54.6.1345-1353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. W., Gross D. C. Physical and functional analyses of the syrA and syrB genes involved in syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1988 Dec;170(12):5680–5688. doi: 10.1128/jb.170.12.5680-5688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaat S. A., van Brussel A. A., Tak T., Pees E., Lugtenberg B. J. Flavonoids induce Rhizobium leguminosarum to produce nodDABC gene-related factors that cause thick, short roots and root hair responses on common vetch. J Bacteriol. 1987 Jul;169(7):3388–3391. doi: 10.1128/jb.169.7.3388-3391.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


