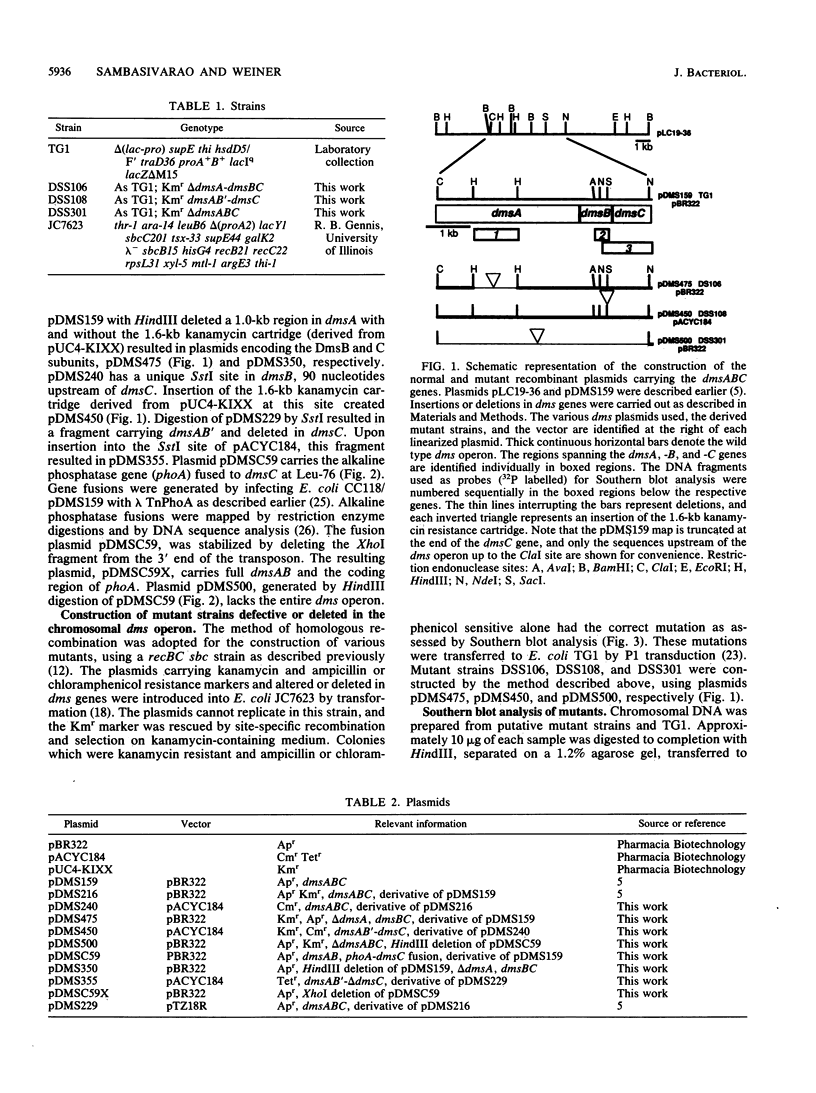
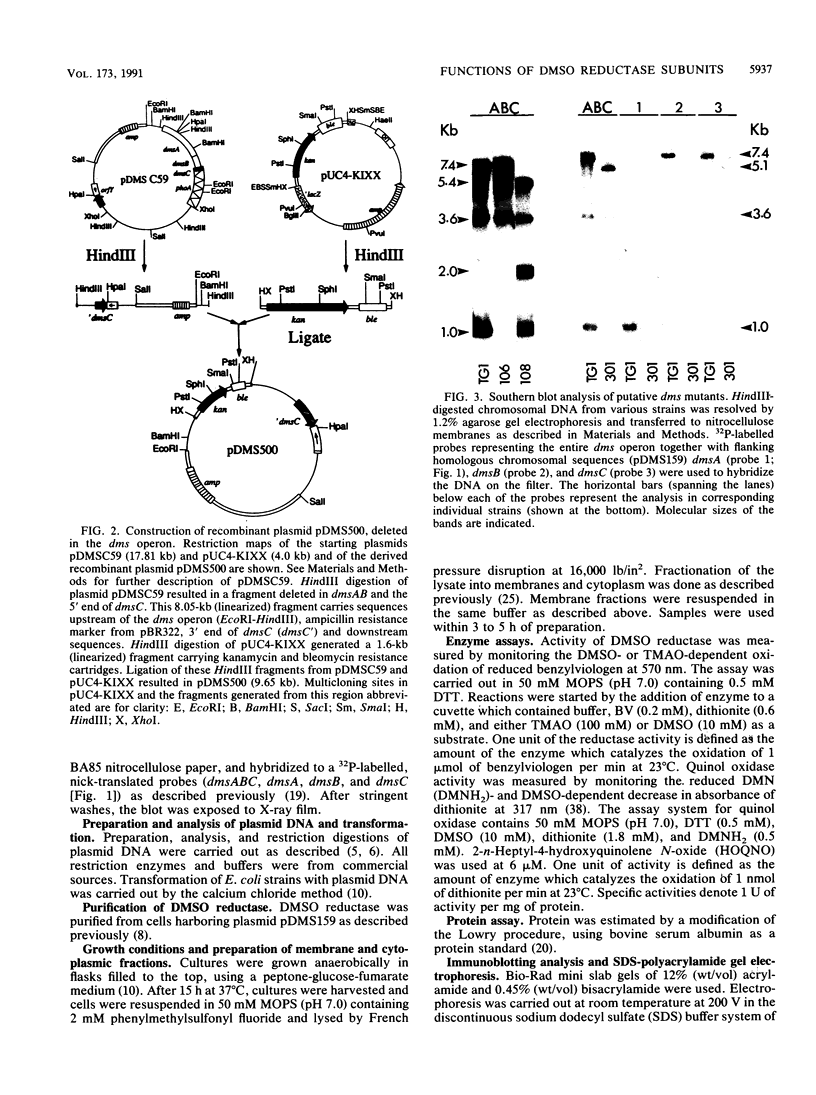
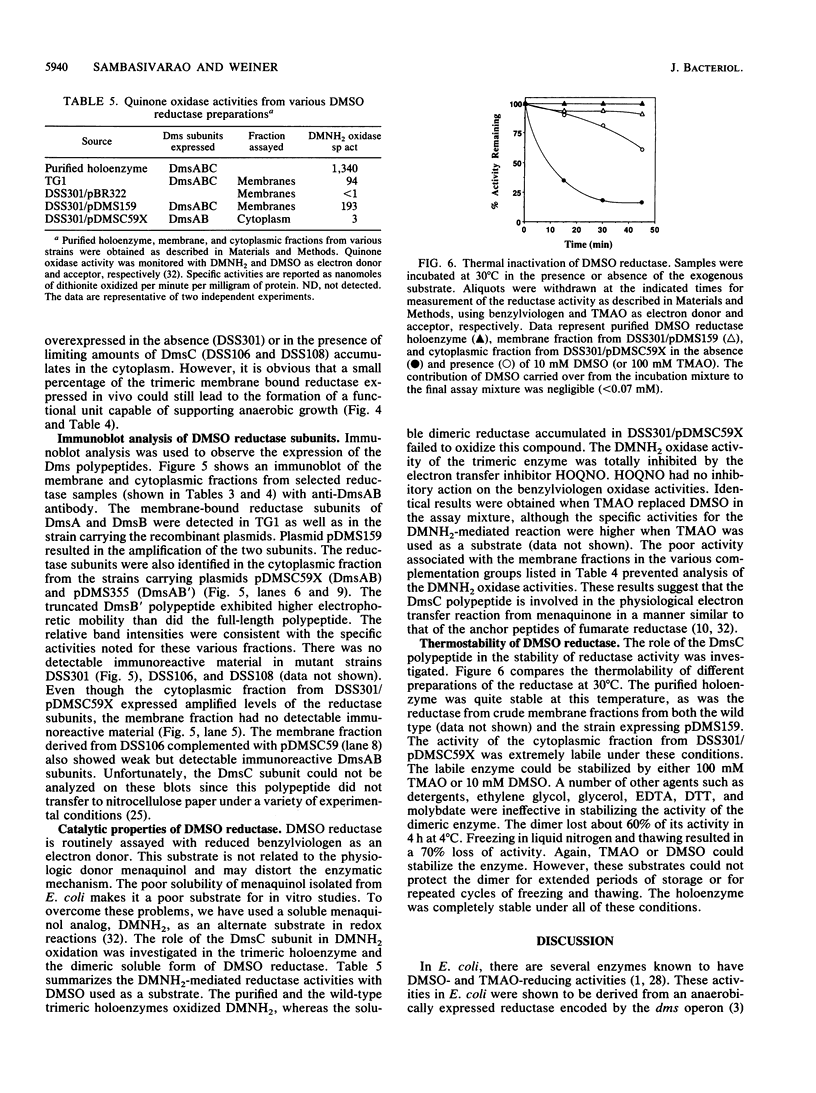
Abstract
Dimethyl sulfoxide (DMSO) reductase of Escherichia coli is a membrane-bound, terminal anaerobic electron transfer enzyme composed of three nonidentical subunits. The DmsAB subunits are hydrophilic and are localized on the cytoplasmic side of the plasma membrane. DmsC is the membrane-intrinsic polypeptide, proposed to anchor the extrinsic subunits. We have constructed a number of strains lacking portions of the chromosomal dmsABC operon. These mutant strains failed to grow anaerobically on glycerol minimal medium with DMSO as the sole terminal oxidant but exhibited normal growth with nitrate, fumarate, and trimethylamine N-oxide, indicating that DMSO reductase is solely responsible for growth on DMSO. In vivo complementation of the mutant with plasmids carrying various dms genes, singly or in combination, revealed that the expression of all three subunits is essential to restore anaerobic growth. Expression of the DmsAB subunits without DmsC results in accumulation of the catalytically active dimer in the cytoplasm. The dimer is thermolabile and catalyzes the reduction of various substrates in the presence of artificial electron donors. Dimethylnaphthoquinol (an analog of the physiological electron donor menaquinone) was oxidized only by the holoenzyme. These results suggest that the membrane-intrinsic subunit is necessary for anchoring, stability, and electron transport. The C-terminal region of DmsB appears to interact with the anchor peptide and facilitates the membrane assembly of the catalytic dimer.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett E. L., Kwan H. S. Bacterial reduction of trimethylamine oxide. Annu Rev Microbiol. 1985;39:131–149. doi: 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- Bilous P. T., Cole S. T., Anderson W. F., Weiner J. H. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol Microbiol. 1988 Nov;2(6):785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Bilous P. T., Weiner J. H. Dimethyl sulfoxide reductase activity by anaerobically grown Escherichia coli HB101. J Bacteriol. 1985 Jun;162(3):1151–1155. doi: 10.1128/jb.162.3.1151-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilous P. T., Weiner J. H. Molecular cloning and expression of the Escherichia coli dimethyl sulfoxide reductase operon. J Bacteriol. 1988 Apr;170(4):1511–1518. doi: 10.1128/jb.170.4.1511-1518.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilous P. T., Weiner J. H. Proton translocation coupled to dimethyl sulfoxide reduction in anaerobically grown Escherichia coli HB101. J Bacteriol. 1985 Jul;163(1):369–375. doi: 10.1128/jb.163.1.369-375.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco F., Iobbi C., Giordano G., Chippaux M., Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol Gen Genet. 1989 Aug;218(2):249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- Cammack R., Weiner J. H. Electron paramagnetic resonance spectroscopic characterization of dimethyl sulfoxide reductase of Escherichia coli. Biochemistry. 1990 Sep 11;29(36):8410–8416. doi: 10.1021/bi00488a030. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Condon C., Lemire B. D., Weiner J. H. Molecular biology, biochemistry and bioenergetics of fumarate reductase, a complex membrane-bound iron-sulfur flavoenzyme of Escherichia coli. Biochim Biophys Acta. 1985 Dec;811(4):381–403. doi: 10.1016/0304-4173(85)90008-4. [DOI] [PubMed] [Google Scholar]
- Condon C., Weiner J. H. Fumarate reductase of Escherichia coli: an investigation of function and assembly using in vivo complementation. Mol Microbiol. 1988 Jan;2(1):43–52. doi: 10.1111/j.1365-2958.1988.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Esposti M. D. Prediction and comparison of the haem-binding sites in membrane haemoproteins. Biochim Biophys Acta. 1989 Dec 7;977(3):249–265. doi: 10.1016/s0005-2728(89)80079-9. [DOI] [PubMed] [Google Scholar]
- Jasin M., Schimmel P. Deletion of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J Bacteriol. 1984 Aug;159(2):783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Latour D. J., Weiner J. H. Assembly of Escherichia coli fumarate reductase holoenzyme. Biochem Cell Biol. 1989 Jun;67(6):251–259. doi: 10.1139/o89-038. [DOI] [PubMed] [Google Scholar]
- Latour D. J., Weiner J. H. Investigation of Escherichia coli fumarate reductase subunit function using transposon Tn5. J Gen Microbiol. 1987 Mar;133(3):597–607. doi: 10.1099/00221287-133-3-597. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Renart J., Reiser J., Stark G. R. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3116–3120. doi: 10.1073/pnas.76.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
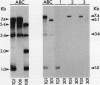
- Sambasivarao D., Scraba D. G., Trieber C., Weiner J. H. Organization of dimethyl sulfoxide reductase in the plasma membrane of Escherichia coli. J Bacteriol. 1990 Oct;172(10):5938–5948. doi: 10.1128/jb.172.10.5938-5948.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Kurihara F. N. Purification and properties of dimethylsulfoxide reductase containing a molybdenum cofactor from a photodenitrifier, Rhodopseudomonas sphaeroides f.s. denitrificans. J Biochem. 1987 Jul;102(1):191–197. doi: 10.1093/oxfordjournals.jbchem.a122032. [DOI] [PubMed] [Google Scholar]
- Silvestro A., Pommier J., Giordano G. The inducible trimethylamine-N-oxide reductase of Escherichia coli K12: biochemical and immunological studies. Biochim Biophys Acta. 1988 Apr 28;954(1):1–13. doi: 10.1016/0167-4838(88)90049-0. [DOI] [PubMed] [Google Scholar]
- Silvestro A., Pommier J., Pascal M. C., Giordano G. The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochim Biophys Acta. 1989 Nov 30;999(2):208–216. doi: 10.1016/0167-4838(89)90220-3. [DOI] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrvold O. B., Strøm A. R. Dimethylsulphoxide and trimethylamine oxide respiration of Proteus vulgaris. Evidence for a common terminal reductase system. Arch Microbiol. 1984 Nov;140(1):74–78. doi: 10.1007/BF00409774. [DOI] [PubMed] [Google Scholar]
- Weiner J. H., Cammack R., Cole S. T., Condon C., Honoré N., Lemire B. D., Shaw G. A mutant of Escherichia coli fumarate reductase decoupled from electron transport. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2056–2060. doi: 10.1073/pnas.83.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. H., MacIsaac D. P., Bishop R. E., Bilous P. T. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J Bacteriol. 1988 Apr;170(4):1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto I., Okubo N., Ishimoto M. Further characterization of trimethylamine N-oxide reductase from Escherichia coli, a molybdoprotein. J Biochem. 1986 Jun;99(6):1773–1779. doi: 10.1093/oxfordjournals.jbchem.a135655. [DOI] [PubMed] [Google Scholar]