Abstract
The 16S rRNA sequences were determined for species of Spirochaeta, Treponema, Borrelia, Leptospira, Leptonema, and Serpula, using a modified Sanger method of direct RNA sequencing. Analysis of aligned 16S rRNA sequences indicated that the spirochetes form a coherent taxon composed of six major clusters or groups. The first group, termed the treponemes, was divided into two subgroups. The first treponeme subgroup consisted of Treponema pallidum, Treponema phagedenis, Treponema denticola, a thermophilic spirochete strain, and two species of Spirochaeta, Spirochaeta zuelzerae and Spirochaeta stenostrepta, with an average interspecies similarity of 89.9%. The second treponeme subgroup contained Treponema bryantii, Treponema pectinovorum, Treponema saccharophilum, Treponema succinifaciens, and rumen strain CA, with an average interspecies similarity of 86.2%. The average interspecies similarity between the two treponeme subgroups was 84.2%. The division of the treponemes into two subgroups was verified by single-base signature analysis. The second spirochete group contained Spirochaeta aurantia, Spirochaeta halophila, Spirochaeta bajacaliforniensis, Spirochaeta litoralis, and Spirochaeta isovalerica, with an average similarity of 87.4%. The Spirochaeta group was related to the treponeme group, with an average similarity of 81.9%. The third spirochete group contained borrelias, including Borrelia burgdorferi, Borrelia anserina, Borrelia hermsii, and a rabbit tick strain. The borrelias formed a tight phylogenetic cluster, with average similarity of 97%. THe borrelia group shared a common branch with the Spirochaeta group and was closer to this group than to the treponemes. A single spirochete strain isolated fromt the shew constituted the fourth group. The fifth group was composed of strains of Serpula (Treponema) hyodysenteriae and Serpula (Treponema) innocens. The two species of this group were closely related, with a similarity of greater than 99%. Leptonema illini, Leptospira biflexa, and Leptospira interrogans formed the sixth and most deeply branching group. The average similarity within this group was 83.2%. This study represents the first demonstration that pathogenic and saprophytic Leptospira species are phylogenetically related. The division of the spirochetes into six major phylogenetic clusters was defined also by sequence signature elements. These signature analyses supported the conclusion that the spirochetes represent a monophylectic bacterial phylum.
Full text
PDF


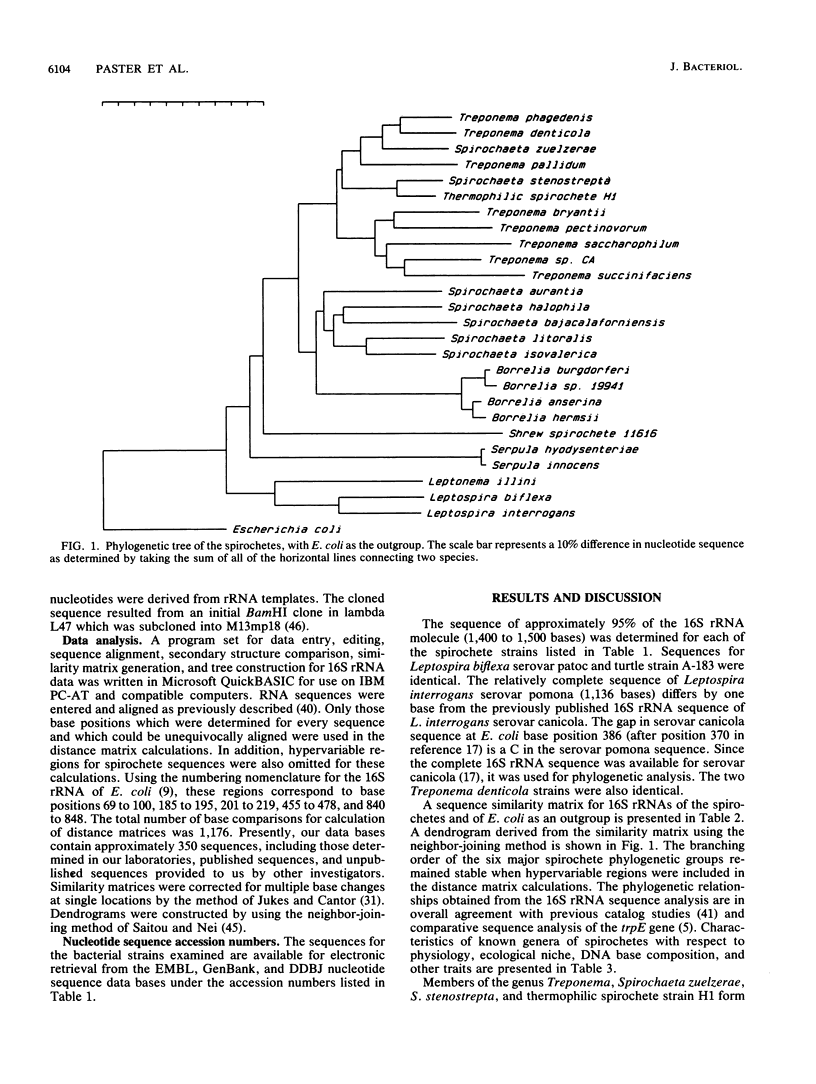
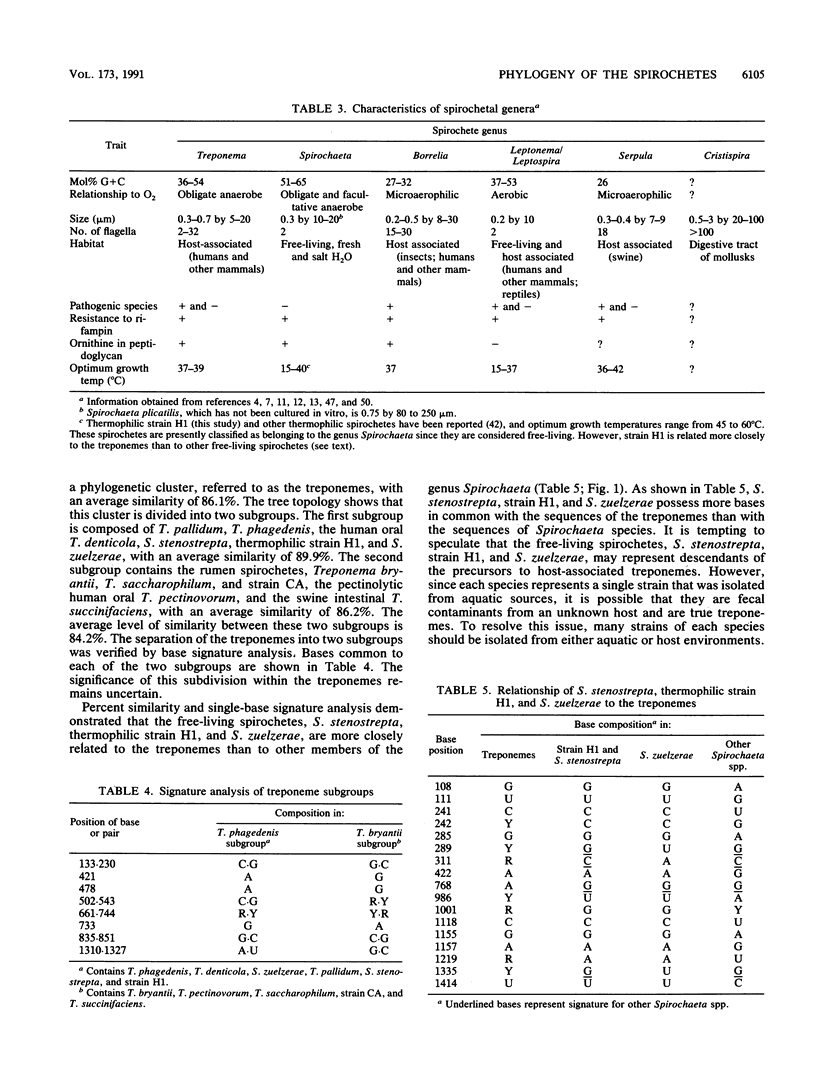




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. F., Johnson R. C., Magnarelli L. A., Hyde F. W., Andreadis T. G. New infectious spirochete isolated from short-tailed shrews and white-footed mice. J Clin Microbiol. 1987 Aug;25(8):1490–1494. doi: 10.1128/jcm.25.8.1490-1494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. F., Magnarelli L. A., LeFebvre R. B., Andreadis T. G., McAninch J. B., Perng G. C., Johnson R. C. Antigenically variable Borrelia burgdorferi isolated from cottontail rabbits and Ixodes dentatus in rural and urban areas. J Clin Microbiol. 1989 Jan;27(1):13–20. doi: 10.1128/jcm.27.1.13-20.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G., Hayes S. F. Biology of Borrelia species. Microbiol Rev. 1986 Dec;50(4):381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahamsha B., Han C. Y., Crawford I. P., Greenberg E. P. Nucleotide sequence and analysis of a gene encoding anthranilate synthase component I in Spirochaeta aurantia. J Bacteriol. 1991 Jan;173(2):541–548. doi: 10.1128/jb.173.2.541-548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Canale-Parola E., Holt S. C., Udris Z. Isolation of free-living, anaerobic spirochetes. Arch Mikrobiol. 1967;59(1):41–48. doi: 10.1007/BF00406315. [DOI] [PubMed] [Google Scholar]
- Canale-Parola E. Physiology and evolution of spirochetes. Bacteriol Rev. 1977 Mar;41(1):181–204. doi: 10.1128/br.41.1.181-204.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaMassa A. J., Adler H. E. Avian spirochetosis: natural transmission by Argas (Persicargas) sanchezi (Ixodoidea: argasidae) and existence of different serologic and immunologic types of Borrelia anserina in the United States. Am J Vet Res. 1979 Jan;40(1):154–157. [PubMed] [Google Scholar]
- Fracek S. P., Jr, Stolz J. F. Spirochaeta bajacaliforniensis sp. n. from a microbial mat community at Laguna Figueroa, Baja California Norte, Mexico. Arch Microbiol. 1985;142:317–325. doi: 10.1007/BF00491897. [DOI] [PubMed] [Google Scholar]
- Fukunaga M., Horie I., Okuzako N., Mifuchi I. Nucleotide sequence of a 16S rRNA gene for Leptospira interrogans serovar canicola strain Moulton. Nucleic Acids Res. 1990 Jan 25;18(2):366–366. doi: 10.1093/nar/18.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg E. P., Canale-Parola E. Spirochaeta halophila sp. n., a facultative anaerobe from a high-salinity pond. Arch Microbiol. 1976 Nov 2;110(23):185–194. doi: 10.1007/BF00690227. [DOI] [PubMed] [Google Scholar]
- Hespell R. B., Canale-Parola E. Amino acid and glucose fermentation by Treponema denticola. Arch Mikrobiol. 1971;78(3):234–251. doi: 10.1007/BF00424897. [DOI] [PubMed] [Google Scholar]
- Hespell R. B., Canale-Parola E. Carbohydrate metabolism in Spirochaeta stenostrepta. J Bacteriol. 1970 Jul;103(1):216–226. doi: 10.1128/jb.103.1.216-226.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978 Mar;42(1):114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde F. W., Johnson R. C. Genetic relationship of lyme disease spirochetes to Borrelia, Treponema, and Leptospira spp. J Clin Microbiol. 1984 Aug;20(2):151–154. doi: 10.1128/jcm.20.2.151-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Harris V. G. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967 Jul;94(1):27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R., Holt S. C., Canale-Parola E. Peptidoglycan of free-living anaerobic spirochetes. J Bacteriol. 1973 Jul;115(1):426–435. doi: 10.1128/jb.115.1.426-435.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. C., Sirdeskmukh R., Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986 Dec;50(4):428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R. M., Fieldsteel A. H., Harris D. L. Genetics of Treponema: characterization of Treponema hyodysenteriae and its relationship to Treponema pallidum. Infect Immun. 1978 Dec;22(3):736–739. doi: 10.1128/iai.22.3.736-739.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., Canale-Parola E. Involvement of periplasmic fibrils in motility of spirochetes. J Bacteriol. 1980 Jan;141(1):359–364. doi: 10.1128/jb.141.1.359-364.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., Canale-Parola E. Physiological diversity of rumen spirochetes. Appl Environ Microbiol. 1982 Mar;43(3):686–693. doi: 10.1128/aem.43.3.686-693.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass P., Jarvis B. D., Corner R. J., Cinco M., Marshall R. B. DNA relatedness among strains of Leptospira biflexa. Int J Syst Bacteriol. 1990 Jul;40(3):231–235. doi: 10.1099/00207713-40-3-231. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmid G. P., Steigerwalt A. G., Johnson S. E., Barbour A. G., Steere A. C., Robinson I. M., Brenner D. J. DNA characterization of the spirochete that causes Lyme disease. J Clin Microbiol. 1984 Aug;20(2):155–158. doi: 10.1128/jcm.20.2.155-158.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Escherichia coli 16S rRNA 3'-end formation requires a distal transfer RNA sequence at a proper distance. EMBO J. 1989 Oct;8(10):3159–3166. doi: 10.1002/j.1460-2075.1989.tb08470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B., Canale-Parola E. Enumeration and selective isolation of rumen spirochetes. Appl Environ Microbiol. 1979 Nov;38(5):965–973. doi: 10.1128/aem.38.5.965-973.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B., Jensen N. S., Casey T. A., Tordoff L. A., Dewhirst F. E., Paster B. J. Reclassification of Treponema hyodysenteriae and Treponema innocens in a new genus, Serpula gen. nov., as Serpula hyodysenteriae comb. nov. and Serpula innocens comb. nov. Int J Syst Bacteriol. 1991 Jan;41(1):50–58. doi: 10.1099/00207713-41-1-50. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VELDKAMP H. Isolation and characteristics of Treponema zuelzerae nov. spec., and anaerobic, free-living spirochete. Antonie Van Leeuwenhoek. 1960;26:103–125. doi: 10.1007/BF02538999. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Hatch T. P., Woese C. R. Eubacterial origin of chlamydiae. J Bacteriol. 1986 Aug;167(2):570–574. doi: 10.1128/jb.167.2.570-574.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]



