Summary
Atherosclerosis is a very complex disease involving both genetic and environmental risk factors, and their interactions. In the general population, genetic polymorphisms of many genes in the pathways of lipid metabolism, inflammation, and thrombogenesis are likely responsible for the wide range of susceptibilities to myocardial infarction, the most deadly consequence of atherosclerosis. To identify these polymorphisms, genetic linkage studies have been carried out in both humans and mouse models. Approximately 40 quantitative trait loci for atherosclerotic disease have been found in humans, and approximately 30 in mice. Recently, genome-wide association studies have been used to identify atherosclerosis-susceptibility polymorphisms. Although finding new atherosclerosis genes through these approaches remains challenging, the pace of finding these polymorphisms is accelerating due to the rapidly improving bioinformatics resources and biotechnologies. The results from these efforts will not only reveal the molecular basis of, but will facilitate finding drug targets and individualized medicine for, atherosclerotic disease.
Introduction
Atherosclerosis is characterized by lipid accumulation, inflammatory response, cell death, and fibrosis in the arterial wall. Our understanding of the pathogenesis of this process has evolved in the last few decades, guiding our efforts to find treatments. Because low-density lipoprotein cholesterol (LDL) is pro-atherogenic, statin drugs were developed to lower plasma LDL cholesterol (LDL-C) levels and reduce the risk of cardiovascular events. Recent studies suggest that reducing LDL-C levels to below current guideline targets further slows down atherogenesis and reduces coronary events (Todd and Farmer, 2006). Statin drugs have reduced new cardiovascular events by one third; although this is significant, it is clear that additional therapies are needed. Based on a plethora of evidence suggesting that increasing plasma high-density lipoprotein cholesterol (HDL-C) levels inhibits atherogenesis, HDL-raising agents are expected to retard atherogenesis. Three classes of HDL-raising agents exist: niacin, fibrate, and statin. Promising new ones include apoA-I-phospholipid complexes, human apoA-I and apoA-I mimetic peptides, and inhibitors of cholesterol ester transfer protein (CETP; questions arise with the failure of Pfizer’s Torcetrapib, though), (Brousseau, 2005). New HDL-raising targets are being sought with genetics and genomics approaches (Rollins et al., 2006; Wang and Paigen, 2005a). The relatively recent appreciation that inflammatory response plays a key role in atherogenesis implies that inhibiting inflammation may provide new anti-atherosclerosis therapy. However, immune response can both promote and retard atherosclerosis (Hansson and Libby, 2006). The challenge will be to inhibit atherosclerosis-specific inflammation without compromising global immune responses.
To better understand the molecular mechanism of atherogenesis, two major approaches have been undertaken. The first is the candidate gene approach, in which genes in known atherogenesis pathways are tested for their role in atherosclerosis in vitro, in vivo and in association studies. This approach has been fruitful and role of many genes in atherogenesis became known as a result (Arnett et al., 2007; Hansson and Libby, 2006; Lusis et al., 2004). The second approach is conducting genome-wide linkage studies to find atherogenesis-regulating quantitative trait loci (QTL). This is an unbiased method that has the potential to find new atherosclerosis genes. Recently, the availability of whole genome sequences in humans and mice, especially the abundant SNP and haplotype information, has made it possible to perform genome-wide association studies, another unbiased approach, to identify disease genes relatively quickly as compared with traditional genetic methods. In deed, the pace of finding disease genes using such unbiased methods has been greatly accelerated in recent years because of the ever improving SNP/haplotype information, experimental design, statistical methods, and the decreasing cost and the high throughput of sequencing. In this review, we focus on finding novel atherosclerosis-regulating genes using genome-wide linkage and genome-wide association studies, summarize successful examples of using these approaches, and present detailed human-mouse comparative genomics maps of all known atherosclerosis-regulating QTL. These maps should be helpful for finding new atherosclerosis-regulating genes.
Genetic linkage studies of atherosclerotic diseases in humans
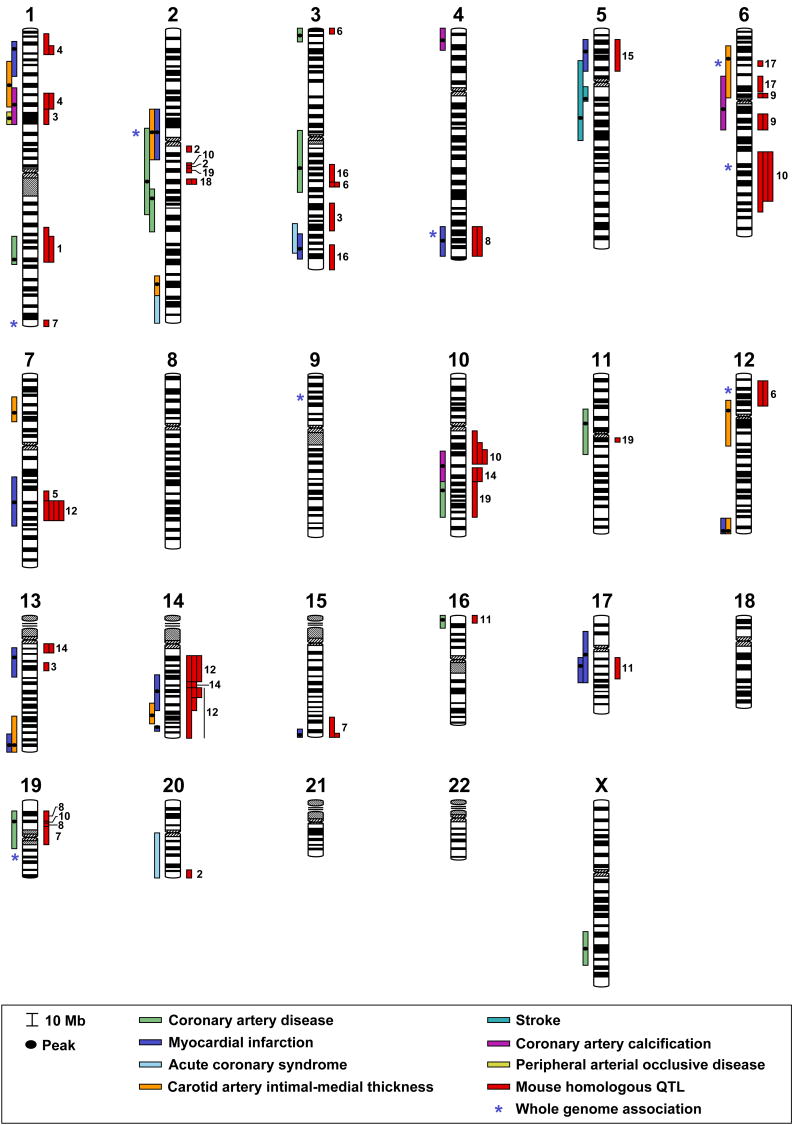
Atherosclerotic disease linkage studies have been conducted in 19 human populations (Table 1). Four of these studies were for linkage to coronary artery disease (CAD), six for linkage to myocardial infarction (MI), one for linkage to both CAD and MI, one for linkage to acute coronary syndrome (ACS), two for linkage to carotid intimal-medial thickness (CIMT), two for linkage to coronary artery calcification (CAC), two for linkage to stroke, and one for linkage to peripheral artery occlusive disease (PAOD). Forty atherosclerotic disease-regulating QTL have been identified: nine for CAD, twelve for MI, one for CAD/MI, three for ACS, nine for CIMT, four for CAC, one for stroke, and one for PAOD. Two meta-analysis of these genome-wide linkage analysis have been reported: in the meta-analysis of four studies, two linkage regions (2q and 3q) in the original studies were confirmed (Chiodini and Lewis, 2003); in another meta-analysis of 5 studies, 4 linkage regions in the original studies (1p, 5p, 12q, 13q) were confirmed, while four new linkage regions that were not found in the original studies (6p, 2 on 8q, 14p) were revealed (Zintzaras and Kitsios, 2006). It is noteworthy that among the 40 human atherosclerosis QTL, only three are replicated in more than one population for the same disease: one CAD QTL on chr 2, one stroke QTL on chr 5, and one MI QTL on chr 17. However, when all the seven disease types in were treated together as “atherosclerotic disease”, then there are 33 QTL regions, and 11 of them are replicates (Fig. 1). This relatively low replicate rate is possibly due to the small sample size and population difference in many studies. By using methods discussed previously (Rollins et al., 2006), we found that 31 of the 40 human QTL are concordant with mouse QTL (Fig. 1), suggesting that identifying the genes underlying the mouse QTL will facilitate identifying the genes underlying the human QTL.
Table 1.
QTLs for atherosclerotic diseases in humans
| Chr | Cytogenetic band Peak (CI)a | Peak cM | Peak (CI) Mbb | Nearest Marker | LOD | Disease | Populationd | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 1p36.13 (p36.21-34.2) | 47 | 21 (13–41) | D1S3669 | 11.7 | MI | U.S., 1613/428 | (Wang et al., 2004) |
| 1p33 (p35.2-31.3) | 76 | 48 (31–61) | D1S2134 | 2.4 | CIMT | U.S., 1242/311 | (Fox et al., 2004) | |
| 1p31.3 (p32.3-31.1) | 92 | 61 (52–80) | … | 2.1 | CAC | U.S. whites, 948/393 sibships | (Turner et al., 2005) | |
| 1p31.1 (p31.1-31.1) | 102 | 71 (70–76) | D1S2895 | 3.9 | PAOD | Icelanders, 116/272 | (Gudmundsson et al., 2002) | |
| 1q31.1 (q24.2-32.1) | 202 | 184 (164–200) | D1S518 | 2.2 | CAD | International, 1168/438 | (Hauser et al., 2004) | |
| 1q44f | 287 | 244 | OR13G1 | OR=1.2 | MI | US whites, 3 case-control studies | (Shiffman et al., 2005) | |
| 2 | 2p11.2 (p13.3-q12.2) | 103 | 85 (70–106) | D2S1790 | 3.3 | MI | U.S., 1613/428 | (Wang et al., 2004) |
| 2p11.2 (p13.3-q12.2) | 103 | 85 (70–106) | D2S1790 | 1.6 | CIMT | U.S., 1242/311 | (Fox et al., 2004) | |
| 2p11.2f | 103 | 86 | rs1010 | OR=1.8 | MI | U.S., 3 case-control studies | (Shiffman et al., 2006) | |
| 2q14.3 (p11.2-q23.3) | 134 | 128 (85–152) | D2S2271 | 2.7 | CAD | U.K.,/1698 | (Samani et al., 2005) | |
| 2q22.1 (q21.2-24.3) | 150 | 142 (134–168) | D2S129 | 3.2 | CAD | Finnish, 364/156 | (Pajukanta et al., 2000) | |
| 2q34 (q33.3-35) | 210 | 214 (205–221) | D2S2944 | 3.1 | CIMT | Mexican Americans, 274/91 | (Wang et al., 2005a) | |
| 2q36-37.3 | 250 | (221–243)e | … | 2.6 | ACS | 61 Australian sib pairs | (Harrap et al., 2002) | |
| 3 | 3p26.1 (p26.3-25.3) | 19 | 6 (0–9) | … | 1.5 | CAD | Europeans, 1464 sibpairs | (Farrall et al., 2006) |
| 3q13.31 (p12.1-q22.1) | 140 | 119 (86–134) | D3S2460 | 3.5 | CAD | International, 1168/438 | (Hauser et al., 2004) | |
| 3q27.2 (q26.31-28) | 199 | 187 (175–191) | D3S1571-3686 | 2.4 | MI | Indo-Mauritians, 535/99 | (Francke et al., 2001) | |
| 3q26-27 | … | (162–190)e | … | 1.8 | ACS | 61 Australian sib pairs | (Harrap et al., 2002) | |
| 4 | 1p16.1 (p16.3-15.33) | 13 | 7 (0–15) | … | 2.0 | CAC | U.S. whites, 948/393 sibships | (Turner et al., 2005) |
| 4q32.3f | 170 | 170 | PALLADIN | OR=1.3 | MI | US whites, 3 case-control studies | (Shiffman et al., 2005) | |
| 4q34.1 (q32.2-35.1) | 176 | 175 (162–185) | D4S2431 | 4.4 | MI | U.S., 1613/428 | (Wang et al., 2004) | |
| 5 | 5p14.3 (p15.2-13.2) | 36 | 22 (10–34) | D5S2845 | 3.7 | MI | U.S., 1613/428 | (Wang et al., 2004) |
| 5q12.1 (q11.2-12.1) | 69 | 59 (57–61) | D5S2080 | 4.4 | Stroke | Icelanders, 914/179 | (Gretarsdottir et al., 2002) | |
| 5q13.3 (p13.3-q14.3) | 82 | 76 (33–83) | D5S424 | 2.0 | Stroke | Northern Swedish, 117/56 | (Nilsson-Ardnor et al., 2005) | |
| 6 | 6p22.3-21.1g | (15–46) | CAD/MI | (Zintzaras and Kitsios, 2006) | ||||
| 6p22.1 (p22.3-12) | 44 | 29 (19–65) | D6S1022 | 2.2 | CIMT | Mexican Americans, 274/91 | (Wang et al., 2005a) | |
| 6p21.33f | 45 | 32 | LTA | OR=1.8 | MI | Japanese, 1133 MI, 1006 controls | (Ozaki et al., 2002) | |
| 6q12 (p21.1-q14.3) | 83 | 72 (42–86) | D6S1053 | 2.2 | CAC | U.S. whites, 94/29 sibships | (Lange et al., 2002) | |
| 6q22.1f | 169 | 118 | ROS1 | OR=1.2 | MI | US whites, 3 case-control studies | (Shiffman et al., 2005) | |
| 7 | 7p14.3 (p15.3-14.1) | 50 | 31 (22–43) | D7S817 | 1.6 | CIMT | U.S., 1242/311 | (Fox et al., 2004) |
| 7q22.2 (q21.12-31.33) | 114 | 104 (87–124) | D7S1799 | 3.6 | MI | U.S., 1613/428 | (Wang et al., 2004) | |
| 8 | 8q13.2-22.2g | (68–99) | CAD/MI | (Zintzaras and Kitsios, 2006) | ||||
| 8q24.21-24.3g | (127–140) | CAD/MI | (Zintzaras and Kitsios, 2006) | |||||
| 9 | Rs10757274 | OR=1.2–1.9 | CAD/MI | 10 different populations | (Helgadottir et al., 2007; McPherson et al., 2007; Wellcome Trust Case Control Consortium, 2007) | |||
| 9p21.3 | 38 | 22 | ||||||
| 10 | 10q22.2 (q21.3-23.2) | 95 | 77 (64–88) | D10S1432 | 3.2 | CAC | U.S. whites, 94/29 sibships | (Lange et al., 2002) |
| 10q23.33 (q23.2-26.11) | 116 | 95 (86–119) | D10S185 | 2.1 | CAD | Indo-Mauritians, 535/99 | (Francke et al., 2001) | |
| 11 | 11p11.2 (p13-q13.1) | 66 | 43 (35–63) | … | 1.7 | CAD | Europeans, 1464 sibpairs | (Farrall et al., 2006) |
| 12 | 12p13.2f | 22 | 11 | TAS2R50 | OR=1.3 | MI | US whites, 3 case-control studies | (Shiffman et al., 2005) |
| 12q12 (p12.1-q13.2) | 56 | 42 (21–55) | D12S1301 | 1.7 | CIMT | U.S., 1242/311 | (Fox et al., 2004) | |
| 12q24.33 (q24.31-24.33) | 161 | 129 (124–132) | D12S1045 | 4.4 | MI | U.S., 1613/428 | (Wang et al., 2004) | |
| 12q24.33 (q24.31-24.33) | 161 | 129 (124–132) | D12S1045 | 4.1 | CIMT | U.S., 1242/311 | (Fox et al., 2004) | |
| 13 | 13q12.3 (q12.12-14.11) | 24 | 30 (23–42) | D13S289 | 2.5 | MI | Icelanders, 713/296 | (Helgadottir et al., 2004) |
| 13q33.3 (q31.1-34) | 94 | 107 (80–109) | D13S796 | 1.3 | CIMT | Mexican Americans, 274/91 | (Wang et al., 2005a) | |
| 13q33.3 (q32.3-34) | 94 | 107 (98–110) | D13S796 | 3.6 | MI | U.S., 1613/428 | (Wang et al., 2004) | |
| 14 | 14p13-q13.1g | (0–32) | CAD/MI | (Zintzaras and Kitsios, 2006) | ||||
| 14q23.1 (q21.3-24.3) | 67 | 60 (49–73) | D14S592 | 4.2 | MI | U.S., 1613/428 | (Wang et al., 2004) | |
| 14q31.2 (q24.2-32.12) | 92 | 83 (70–92) | D14S606 | 1.8 | CIMT | U.S., 1242/311 | (Fox et al., 2004) | |
| 14q32.2 (q32.2-32.31) | 126 | 100 (98–101) | D14S1426 | 3.9 | MI | Germans, 1406/513 | (Broeckel et al., 2002) | |
| 15 | 15q26.3 (q26.2-26.3) | 113 | 97 (96–99) | D15S120 | 4.2 | CAD/MI | U.S., 21/1 | (Wang et al., 2003a) |
| 16 | 16p13.3 (p13.3-13.2) | 9 | 4 (0–8) | D16S3027 | 3.1 | CAD | Indo-Mauritians, 535/99 | (Francke et al., 2001) |
| 17 | 17q11.2 (p12-q22) | 51 | 23 (14–50) | … | 2.9 | MI | Europeans, 739 sibpairs | (Farrall et al., 2006) |
| 17q21.2 (q12-22) | 67 | 36 (32–50) | … | 1.6 | MI | Europeans, 344 sibpairs | (Farrall et al., 2006) | |
| 19 | 19p13.12 (p13.2-q13.11) | 44 | 16 (9–40) | D19S252 | 1.7 | CAD | International, 1168/438 | (Hauser et al., 2004) |
| 19q13.2f | 65 | 47 | Rs11881940 | OR=1.9 | MI | U.S., 3 case-control studies | (Shiffman et al., 2006) | |
| 20 | 20q11-13 | … | (28–62)e | … | 1.6 | ACS | 61 Australian sib pairs | (Harrap et al., 2002) |
| X | Xq25 (q23-26) | 82 | 129 (109–137) | DX1047 | 3.5 | CAD | Finnish, 364/156 | (Pajukanta et al., 2000) |
Peak cytogenetic bands were retrieved from Ensembl Human NCBI 35, version 37.35j (accessed 3-1-06), according to the positions of the peak markers. Confidence intervals were determined either by the 1.5-LOD drop method when LOD score curves and markers were available, found in LOD score tables, or by using ± 15 cM around the peak marker in the absence of the above information.
Mb: based on Ensembl Human NCBI 35, version 37.35j (accessed 3-1-06).
Mouse homologous chromosome regions of the human QTLs were determined as described in the text. Peak position is listed first, followed by regions arranged by chromosome number.
Number of individuals/families.
Intervals estimated according to positions of cytogenetic bands.
Detected in genome-wide association studies.
Results from Meta-analysis
ACS, acute coronary syndrome (MI+unstable angina); CAC, coronary artery calcification; CAD, coronary artery disease; CIMT, carotid artery intimal-medial thickness; MI, myocardial infarction; PAOD, peripheral arterial occlusive disease.
Figure 1.
Chromosome map of human and mouse concordant QTLs for atherosclerotic diseases. Human HDL-C QTLs are represented by bars to the left of each chromosome. Each bar represents a QTL from one population as shown in Table 1. QTL sizes are given either as 1.5-LOD drop intervals (if LOD score figures are available), or ± 15 cM centered around the LOD score peak (when LOD score figures are unavailable). Human homologues of mouse HDL-C QTLs are represented by red bars to the right of each chromosome, and the chromosome numbers of these mouse QTLs are to their right.
Putative disease genes have been found from only three QTL for atherosclerotic disease, but the pace of identifying these genes is expected to be accelerating (Farrall et al., 2006; Hakonarson et al., 2005). The DeCode Genetics group in Iceland mapped a QTL for both MI and stroke to chr 13q12-13, and subsequently identified 5-lipoxygenase activating protein (ALOX5AP, also called FLAP) as the underlying gene (Helgadottir et al., 2004). ALOX5AP contributes to the conversion of ALOX5 in the leukotriene pathway, yielding to production of leukotriene B4, one of the most potent leukocyte pro-inflammatory mediators. This finding is consistent with the discovery that 5-lipoxygenase (Alox5) is the underlying gene for an atherosclerosis QTL on mouse Chr 6, and that ALOX5/ALOX5AP pathway influences the development of atherosclerosis (discussed below). In another study, Wang et al. studied a large family with 13 patients who displayed an autosomal dominant pattern of CAD (nine of whom developed acute MI). They found a linkage of CAD/MI to a region on 15q26, which consists of 93 genes. One of the genes, myocyte-enhancing factor 2A (MEF2A), is expressed in endothelial cells of coronary arteries. A 21-base pair deletion was identified in exon 11 in all ten living members with CAD in the family, and not in family members and an additional 119 individuals with normal angiograms, strongly suggesting that the deletion is responsible for CAD and MI in this large family (Wang et al., 2003a). MEF2A mutations may be a rare cause of CAD and MI because they are present in less than 2% of a U.S. population of 207 CAD/MI patients (Bhagavatula et al., 2004). Later, in another two studies, the 21-base pair deletion was found in a total of six subjects (3 in each study) older than 60 years of age who had no history nor symptoms of CAD (Kajimoto et al., 2005b; Weng et al., 2005), which seems to argue against the causality of the 21-base pair deletion in CAD/MI (Altshuler and Hirschhorn, 2005). However, in these later two studies, the CAD status in control subjects was not confirmed with angiography, as Wang et al did in the original publication (Wang et al., 2003a), making the conclusion not definitive. A recent study found that a P279L variant in MEF2A is associated with the risk of MI (11 of 483, or 2.3%, in MI patients, and 9 of 1189, or 0.8%, in controls; OR=3.06) (Gonzalez et al., 2006). In this study, the CAD status in controls was not confirmed with angiography either; however, this might add to the strength of the association because some of the nine “controls”, who were all under 50 years of age, may develop MI at older ages. More recently, association studies were done on candidate genes in the 1-LOD supporting interval of the 3q13 QTL for CAD they identified (Hauser et al., 2004), and authors found that four closely located genes on Chr 3 were associated with the risk for CAD: CDGAP at 120.5 Mb, MYLK at 124.8 Mb, and KALRN at 125.3 Mb in one study, (Wang et al., 2007), and GATA2 at 129.7 Mb in another study (Connelly et al., 2006). The first three are all in the GTPase-signaling pathway. How these genes affect CAD risk is not clear, although KALRN inhibit inducible nitric oxide synthase, and GATA1 may affect endothelial functions.
Genetic linkage studies of atherosclerosis in mice
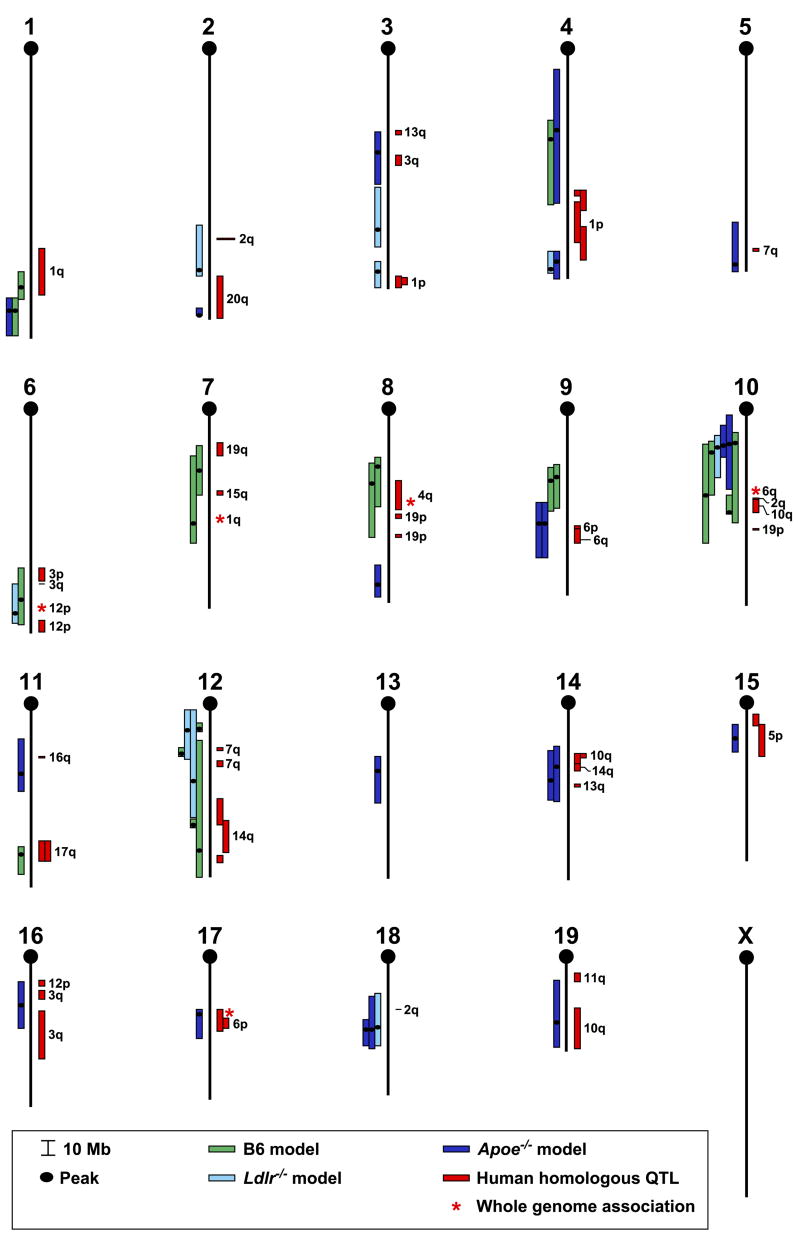
The three most often used mouse models of atherosclerosis are the following: 1) a high fat model in which inbred mice are fed a high fat and cholesterol diet (15% fat, 1.25% cholesterol, and 0.5% cholic acid), 2) Apoe-deficient mice fed either chow, a Western diet (21% fat and 0.15% cholesterol), or a high fat and cholesterol diet without cholic acid, and 3) Ldlr-deficient mice fed either a Western diet or a high fat and cholesterol diet. The latter two models are also called sensitized models (Dansky et al., 2002). Some early QTL studies took advantages of the existing recombinant inbred lines, such as BxH and CxB (Paigen et al., 1987a; Paigen et al., 1987b; Paigen et al., 1987c). Since then, fourteen different F2 or N2 crosses have been generated to find atherosclerosis-regulating QTL: seven crosses involved normal inbred mice, five involved inbred mice with Apoe-deficiency, and two involved inbred mice with Ldlr-deficiency (Table 2). Thirty different mouse atherosclerosis QTL have been found: fifteen in one cross, eleven in two different crosses, and four in three or more crosses. Among the thirty atherosclerosis-regulating QTL, seven were found only in the high fat model, twelve only in the Apoe-deficient model, four only in the Ldlr-deficient model, two in both the Apoe-deficient and the Ldlr-deficient models, and five in both the high fat and the sensitized models. Among the 30 QTL, 13 are replicates (the same QTL region found in different studies). A comparative map (Fig. 2) of mouse and human atherosclerosis-regulating QTLs indicates that most of the mouse QTL found in both the high fat model (86%) and the sensitized models (61%) are concordant with the human QTL. In all, twenty of the thirty mouse QTLs are concordant with the human QTL. This comparative QTL map will be helpful in two ways. First, it will help researchers focus on the concordant QTL because finding the underlying genes in the mouse might facilitate finding them in humans. Second, by overlapping the concordant QTL and excluding the non-overlapping regions, based on the assumption that the same orthologous genes underlie the QTL in both species, we might be able to significantly narrow many of the mouse QTL. Of course, this assumption may not always be true. Mouse QTL can also be narrowed with other bioinformatics methods, such as combined cross analysis, haplotype analysis, and whole-genome association studies (DiPetrillo et al., 2005). Candidate genes can be analyzed for sequence and expression patterns, and a shortened list of candidates can be verified in animal models and in human association studies (Wang et al., 2005b).
Table 2.
Quantitative trait loci for atherosclerosis in inbred mouse crosses
| Chr | Crossa | markerb | cM (CI)c | Mb (CI)d | LODe | locus | Reference |
|---|---|---|---|---|---|---|---|
| 1 | B × H and C × B RI lines | D1Mit159 | 82 | 162 (151–170) | … | Ath1 | (Paigen et al., 1987c; Phelan et al., 2002) |
| (B6 × A) F2 | 1–169 Mb | 100 | 178 (169–195) | 3.4 | Ishimori et al, unpublished | ||
| (B6.129-Apoe−/− × FVB.129-Apoe−/−) F2 | D1Mit359 | 100 | 178(169–195) | 3.3 | Ath9 | (Dansky et al., 2002) | |
| 2 | (PERA × B6.129-Ldlr−/−) × B6.129-Ldlr−/− | D2Mit405 | 69 | 148 (117–152) | 2.8 | Athla1 | (Bretschger Seidelmann et al., 2005) |
| (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13476938 | 107 | 179 (174–179) | 3.3 | Ath28 | (Smith et al., 2006) | |
| 3 | (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13477166 | 33 | 67 (53–89) | 2.7 | Ath23 | (Smith et al., 2006) |
| (B6.129-Ldlr−/− × FVB.129-Ldlr −/−) F2 | D3Mit57 | 55 | 119 (90–131) | 4.1 | Ascla4 | (Teupser et al., 2006) | |
| (FVB.129-Ldlr −/− × B6.129-Ldlr −/−) F2 | D3Mit45 | 79 | 148 (141–160) | 3.3 | Ascla3 | (Teupser et al., 2006) | |
| 4 | (B6.129-Apoe−/− × C3H.129-Apoe−/−) F2 | D4Mit111 | 22 (3–50) | 52 (10–102) | 3.0 | (Wang et al., 2003b) | |
| (SM × NZB) F2 | D4Mit44 | 29 (18–51) | 59 (46–104) | 3.6 | Ath8 | (Korstanje et al., 2004) | |
| (A × B6.129-Apoe−/−) F2 | 4–135 Mb | 76 | 143 (136–154) | 4.6 | Ishimori et al, unpublished | ||
| (MOLF × B6-Ldlr−/−) × B6-Ldlr−/− | D4Mit127 | 78 (66–81) | 149 (136–151) | 6.2 | Athsq1 | (Welch et al., 2001) | |
| 5 | (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13478585 | 84 | 147 (130–139) | 3.4 | Ath24 | (Smith et al., 2006) |
| 6 | (CAST × B6) F2 | D6Mit256 | 61 (46–71) | 127 (105–144) | 6.7 | Artles | (Mehrabian et al., 2001) |
| (MOLF × B6-Ldlr−/−) × B6-Ldlr−/− | D6Mit110 | 64 (52–70) | 136 (115–143) | 6.7 | Athsq2 | (Welch et al., 2001) | |
| 7 | (B6 × DBA/2) F2 | D7Mit193 | 25 | 51 (33–68) | 3.7 | Aorls2 | (Colinayo et al., 2003) |
| A × B and B × A RI lines | Tyr | 44 (20–50) | 87 (41–100) | … | Ath3 | (Stewart-Phillips et al., 1989) | |
| 8 | (B6 × A) F2 | 8–54 Mb | 22 | 36 (31–67) | 2.8 | Ishimori et al, unpublished | |
| (B6 × DBA/2) F2 | D8Mit41 | 29 | 49 (39–88) | 3.4 | (Colinayo et al., 2003) | ||
| (A × B6.129-Apoe−/−) F2 | 8–109 Mb | 60 | 120 (106–129) | 2.0 | Ishimori et al, unpublished | ||
| 9 | (B6 × A) F2 | 9–48 Mb | 25 | 43 (34–63) | 4.1 | Ishimori et al, unpublished | |
| SWR (SWR × SJL) | D9Mit330 | 26 | 45 (36–66) | Svenson et al, unpublished | |||
| (A × B6.129-Apoe−/−) F2 | 9–64 Mb | 42 | 74 (60–98) | 3.7 | Ishimori et al, unpublished | ||
| (B6.129-Apoe−/− × C3H.129-Apoe−/−) F2 | D9Mit156 | 42 | 74 (60–98) | 4.1 | Ath22 | (Su et al., 2006) | |
| 10 | (B6 × 129) F2 | D10Mit213 | 10 (5–40) | 19 (12–74) | … | Ath20g | (Ishimori et al., 2004) |
| (B6.129-Apoe−/− × FVB.129-Apoe−/−) F2 | D10Mit213 | 11 (0–29) | 20 (0–51) | 5.1 | Ath11 | (Dansky et al., 2002) | |
| (A × B6.129-Apoe−/−) F2 | 10–27 Mb | 12 | 22 (7–29) | 2.8 | Ishimori et al, unpublished | ||
| (B6.129-Ldlr −/− × FVB.129-Ldlr −/−) RpF2 | D10Mit16 | 16 | 21 (14–43) | 13.1 | Ascla1 | (Teupser et al., 2006) | |
| (B6 × A) F2 | 10–27 Mb | 20 | 26 (18–55) | 3.9 | Ishimori et al, unpublished | ||
| (B6 × DBA/2) F2 | D10Mit42 | 30 (14–45) | 55 (20–88) | 4.5 | Aorls1 | (Colinayo et al., 2003) | |
| (B6 × 129) F2 | D10Mit31 | 34 (30–36) | 67 (55–68) | 6.6 | Ath17 | (Ishimori et al., 2004) | |
| 11 | (B6.129-Apoe −/− × C3H.129-Apoe −/−) F2 | D11Mit236 | 20 | 44 (20–56) | 2.4 | (Su et al., 2006) | |
| (B6 × 129) F2 | D11Mit333 | 60 (55–70) | 99 (94–113) | … | Ath19g | (Ishimori et al., 2004) | |
| 12 | (B6-db/db × BKS) F2 | D12Mit49 | 3 (2–4) | 13 (9–15) | 2.5 | Ath6 | (Mu et al., 1999) |
| (B6.129-Ldlr−/− × FVB.129-Ldlr−/−) RpF2 | D12Mit82 | 3 | 13 (0–39) | 3.9 | Ascla5 | (Teupser et al., 2006) | |
| (B6 × 129) F2 | D12Mit243 | 16 (13–17) | 36 (32–38) | 3.7 | Ath18 | (Ishimori et al., 2004) | |
| (B6.129-Ldlr−/− × FVB.129-Ldlr−/−) RpF2 | D12Mit189 | 24 (0–34) | 55 (0–79) | 4.8 | Ascla6 | (Teupser et al., 2006) | |
| SWR × (SWR × SJL), SWR × SJL RI | D12Mit158 | 38 (35–40) | 85 (81–86) | Ath7 | Svenson et al, unpublished | ||
| (B6 × 129) F2 | D12Mit7 | 50 (10–70) | 103 (27–119) | … | Ath21g | (Ishimori et al., 2004) | |
| 13 | (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13481782 | 26 | 43 (33–67) | 2.8 | Ath25 | (Smith et al., 2006) |
| 14 | (B6.129-Apoe−/− × FVB.129-Apoe−/−) F2 | D14Mit60 | 15 (10–40) | 45 (28–70) | 2.5 | Ath13 | (Dansky et al., 2002) |
| (A × B6.129-Apoe−/−) F2 | 14–52 Mb | 22 | 55 (31–69) | 2.6 | Ishimori et al, unpublished | ||
| 15 | (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13482467 | 14 | 20 (10–29) | 3.3 | Ath22 | (Smith et al., 2006) |
| 16 | (B6.129-Apoe−/− × FVB.129-Apoe−/−) F2 | D16Mit103 | 22 | 29 (13–45) | 2.5 | (Dansky et al., 2002) | |
| 17 | (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13482966 | 20 | 36 (33–54) | 4.3 | Ath26 | (Smith et al., 2006) |
| 18 | (B6.129-Ldlr−/− × FVB.129-Ldlr−/−) F2 | D18Mit23 | 21 | 44 (21–57) | 3.1 | (Teupser and Breslow, 2004) | |
| (A × B6.129-Apoe−/−) F2 | 18–52 Mb | 22 | 46 (23–59) | 2.5 | Ishimori et al, unpublished | ||
| (AKR.129-Apoe−/− × DBA/2.129-Apoe−/−) RpF2 | rs13483316 | 22 | 41 (39–57) | 3.6 | Ath27 | (Smith et al., 2006) | |
| 19 | (B6.129-Apoe−/− × FVB.129-Apoe−/−) F2 | D19Mit120 | 41 (6–54) | 42 (12–58) | 3.8 | Ath16 | (Dansky et al., 2002) |
| ? | A × B RI, B × A RI | … | Ath2 | (Paigen, 1995) |
RI, recombinant inbred strains; F2, intercross; RpF2, reciprocal F2—both strains were used as female and male parental strains, and the F2s were combined for analysis.
Peak marker. The non-MIT markers are all SNP markers with their positions (for example, 1–169 Mb means a SNP on Chr 1, at 169 Mb).
cM, peak centimorgan. Genetic distances were retrieved from MGI at http://www.informatics.jax.org. CI: 95% confidence interval.
Mb, megabase, derived from the physical positions (Ensembl NCBI m36, Gene build April 2006) of the markers at the corresponding cM position. In the absence of information on CI in cM, peak ± 10 cM is used instead.
LOD, logarithm of the odds ratio
Figure 2.
Chromosome map of mouse and human concordant atherosclerosis QTLs. A vertical black line represents each chromosome, with the centromere at the top. Genetic distances in Mb from the centromere are shown by the scale at the lower left of the figure. Mouse atherosclerosis QTLs are represented by bars to the left of each chromosome. Each bar represents a QTL from one cross as shown in Table 2. QTL sizes are given either as 95% confidence intervals (CIs), 1.5-LOD drop intervals (if 95% CIs are not available but LOD score figures are available), or ± 10 cM centered around the LOD score peak (when neither CIs nor LOD score figures are available). To change cM into Mb, we found MIT markers at the cM positions in MGI (informatics.jax.org) and retrieved their Mb positions in Ensembl Mouse Genome Server NCBI m36. Mouse homologues of human HDL-C QTLs are represented by red bars to the right of each chromosome, and the chromosome numbers of these human QTLs are to their right.
Two of the genes underlying mouse atherosclerosis QTL have been identified: Alox5 for the QTL Artles on Chr 6, and Tnfsf4 (Ox40l) for the QTL Ath1 (atherosclerosis susceptibility 1) on Chr 1 (Table 2). Artles was identified in a cross between CAST and C57BL/6 (Mehrabian et al., 2001). By using Alox5 targeted mutant mice with or without Ldlr-deficiency, a positional candidate, 5-lipoxygenase (Alox5), was shown to increase atherosclerosis (Ghazalpour et al., 2006; Mehrabian et al., 2002). Other evidence that the ALOX5/ALOX5AP pathway is involved in atherogenesis seems compelling: genetic linkage and association studies in humans have revealed that ALOX5AP confers risk of MI and/or stroke (Helgadottir et al., 2005; Helgadottir et al., 2004; Kajimoto et al., 2005a; Kaushal et al., 2007; Lohmussaar et al., 2005); promoter variants of ALOX5 are associated with CIMT in humans (Dwyer et al., 2004); expression levels of ALOX5 are elevated in symptomatic plaques and are associated with acute ischemic syndromes (Cipollone et al., 2003; Qiu et al., 2006); ALOX5AP inhibitor reduces atherosclerosis in mice (Jawien et al., 2006). Because ALOX5 and ALOX5AP are involved in leukotriene formation, leukotriene receptors and leukotriene-forming enzymes are studied for their role in atherosclerosis. It was found that CP-105,696, a leukotriene B4 (LTB4) receptor antagonist, decreases atherosclerosis in both Apoe−/− and Ldlr−/− mouse models, and the effect was dependent on the presence of CCL2 (MCP1) because the drug has no effect in Ccl2−/− mice (Aiello et al., 2002). In addition, targeted mutation of LTB4 receptor 1 (Ltb4r1) leads to reduced early atherosclerosis (Heller et al., 2005; Subbarao et al., 2004). Furthermore, a polymorphism of leukotriene A4 hydrolase (LTA4H), whose gene product hydrolyzes LTA4 into the leukocyte chemoattractant LTB4, is associated with the risk of MI in an Icelandic cohort (Helgadottir et al., 2004). The above observations strongly suggest that ALOX5/ALOX5AP pathway influences the progress of atherosclerotic disease.
However, it is noteworthy that conflicting data do exist. In both Apoe−/− and Ldlr−/− mouse models, ALOX5 deficiency does not significantly affect lesion development in a study by Zhao et al (Zhao et al., 2004), in contrast to the observation by Mehrabian et al (Ghazalpour et al., 2006; Mehrabian et al., 2002). Apart from the differences in study design (age of the mice, length of diet, methods for measuring the lesions), the Alox5 knockout mice used by these two different groups likely had different lengths of 129 flanking regions, which may exert additional effects on atherogenesis other than those by Alox5 itself. In fact, the Alox5 region contains at least two, and may be as many as four, genes that affect atherosclerosis (Ghazalpour et al., 2006). In addition, the association of ALOX5AP polymorphisms with MI and/or ischemic stroke has not been confirmed in other studies (Koch et al., 2007; Meschia et al., 2005; Zee et al., 2006). This is most likely due to the population/ethnic differences, as the associations are more significant in Icelandic, less or not so in non-Icelandic populations. Therefore, ALOX5/ALOX5AP pathway may confer ethnicity-specific risk for MI and stroke. Nonetheless, the use of the relatively isolated Icelandic population reveals that ALOX5/ALOX5AP pathway influences atherosclerotic disease. One possible mechanism is that macrophage ALOX5 cascade generates leukotrienes, which then act on neighboring endothelial cells (and possibly also T cells and macrophages themselves), thereby activating the release of CC and CXC chemokines, resulting in the recruitment of inflammatory cells into the artery (Zhao et al., 2004). Therefore ALOX5 and ALOX5AP may be targeted for treating human atherosclerotic disease.
The second atherosclerosis-regulating QTL gene found in the mouse was Tnfsf4 (OX40 ligand). OX40 ligand and its receptor OX40 belong to the TNF/TNFR superfamily. OX40 is mainly expressed on activated T cells, and OX40L is expressed on antigen-presenting cells (B cells, dendritic cells, macrophages, and endothelial cells) and T cells. The function of the OX40-OX40L costimulatory pathway has been studied in Ox40- (Kopf et al., 1999) and Ox40l-deficient mice (Chen et al., 1999; Murata et al., 2000). These studies demonstrated that OX40-OX40L interactions are crucial for optimum T-cell function and the generation of memory T cells by promoting the survival of effector T cells after antigen priming (Sugamura et al., 2004). Because Ox40l is a candidate gene for Ath1 (Paigen et al., 1987c; Phelan et al., 2002), we studied the role of the OX40/OX40L pathway in atherosclerosis. We found that OX40L is expressed on endothelial cells, macrophages, and lymphocytes in mouse atherosclerotic lesions. OX40L is pro-atherogenic in mice: Ox40l-deficient mice have smaller lesions, and transgenic mice over-expressing Ox40l have larger lesions than do controls (Wang et al., 2005c). More recently, van Wanrooij et al found that treating Ldlr−/− mice with antibody again OX40 reduces atherosclerosis by 53% (van Wanrooij et al., 2007), supporting the pro-atherogenic role of OX40L. Based on the mouse studies, we performed candidate gene association studies and found that OX40L may also influence atherosclerosis in humans: in two independent populations, the less common allele of SNP rs3850641 in OX40L is significantly more frequent (P ≤ 0.05) in individuals with myocardial infarction than it is in controls (Wang et al., 2005c). More recently, polymorphisms of OX40 were found to be associated with the risk of MI (Ria et al., 2006), confirming that OX40L/OX40 pathway influences the risk of MI. However, the underlying mechanism is unclear. The OX40L expressed on vascular ECs might bind to OX40 on lymphocytes, facilitating their activation and recruitment into the arterial wall. Or, by reverse signaling, ligation of OX40L on vascular ECs, macrophages, and lymphocytes may induce the production of chemoattractants, drawing and directing leukocytes from the blood into atherosclerotic plaques (Wang, 2006). This possibility is supported by the observation that interaction of recombinant OX40 with human umbilical vein endothelial cells (HUVECs) induces the endothelial cells to produce the CC chemokine RANTES (CCL5) (Kotani et al., 2002), a chemoattractant for monocyte and T cells. RANTES may promote atherosclerosis: intraperitoneal injection of RANTES receptor antagonist Met-RANTES reduces the progression of atherosclerosis in Ldlr−/− mice, accompanied by decreased leukocyte infiltration into lesions (Veillard et al., 2004). It is also possible that the engagement of OX40L and OX40 may induce the production of inflammatory cytokines, as shown in a recent study indicating that the engagement of OX40L on human airway SMCs with recombinant OX40 releases IL-6, a pro-inflammatory cytokine, from these SMCs (Burgess et al., 2004).
There is evidence that angiopoietin-like 3 (Angptl3) underlies Ath8, an atherosclerosis-regulating QTL on mouse Chr 4 (Korstanje et al., 2004): Angptl3 has three non-synonymous polymorphisms between NZB and SM, the parental strains of the cross revealing Ath8; ANGPTL3 inhibits lipoprotein lipase activity and increases plasma total cholesterol and triglycerides levels (Koster et al., 2005); ANGPTL3 binds to integrin alpha(v)beta(3), suggesting it plays a role in angiogenesis and atherogenesis (Camenisch et al., 2002); polymorphisms of ANGPTL3 are associated with the size of coronary atherosclerotic plaques in one human population (Korstanje et al., 2004).
Genome-wide association studies
Testing the association of a gene with atherosclerosis disease is usually motivated by existing pieces of evidence that the gene might be involved in certain atherogenic pathways. However, single gene association studies are notorious for high incidence of false positive findings and low level of reproducibility. Recently, the advent of dense SNPs, detailed haplotype information and low cost and high throughput genotyping has made it possible to carry out unbiased whole genome association studies to find disease genes, including those for atherosclerosis. In a pioneering, large-scale, case-control association study of a Japanese population, Ozaki et al used 92,788 gene-based single-nucleotide polymorphism (SNP) markers across the genome and identified the association of LTA (lymphotoxin-alpha) polymorphisms with susceptibility to MI (Ozaki et al., 2002). This association with CAD/MI has been replicated in some (Laxton et al., 2005; Mizuno et al., 2006; PROCARDIS-Consortium, 2004) but not other studied populations (Clarke et al., 2006), suggesting a population-dependent effect. Following the results of their initial study, Ozaki et al. used an E. coli two-hybrid system and phage display to show that the LTA protein binds to LGALS2 (lectin, galactose-binding, soluble 2). They then found that an SNP in LGALS2 is significantly associated with susceptibility to MI in a Japanese population, and that smooth muscle cells and macrophages in the atherosclerotic lesions express both LGALS2 and LTA (Ozaki et al., 2004). However, the association of LGALS2 polymorphisms with MI was not confirmed in a Caucasian population (Mangino et al., 2006), which may be due to population heterogeneity. More studies are needed to address this inconsistency. More recently, prompted by the finding that binding of LTA to its receptor strongly activates NF-κB, an important regulator of pro- and anti-inflammatory genes and cell survival in atherosclerotic lesions (de Winther et al., 2005; Monaco and Paleolog, 2004), by proteasomal degradation of its inhibitory partner, IκB19, Ozaki et al tested the association of a variation in the genes encoding proteasomal proteins and the risk of MI (Mizuno et al., 2006). The study revealed that a common SNP (rs1048990) in the proteasome subunit alpha type 6 gene (PSMA6) confers risk of MI. The association was confirmed in a second cohort of 867 MI and 1,104 control subjects. They further found that rs1048990 affects the transcription activity of PSMA6, and suppression of PSMA6 expression using siRNA reduces the activation of NF-κB. These results demonstrate that large-scale association studies can quickly identify novel disease-causing genes (such as LTA), and, in combination with functional genomics studies, additional disease genes can be identified (for example PSMA6). Later, in a large-scale association studies using 11,053 SNPs, Shiffman et al found four gene variants associated MI (PALLD, KOS1, TAS2R50, OR13G1), and the association was confirmed in two additional populations (Shiffman et al., 2005). In another large scale association study using 11,647 SNPs, these authors found that VAMP8 and HNRPUL1 were associated with the risk of MI, and the association was confirmed in two additional populations (Shiffman et al., 2006). The possible functions of these two genes in MI are not clear; VAMP7 may activate platelet thereby affecting MI. more recently, three different studies, in which 100K- to 500K-SNP sets and 12 different populations were used, identified SNPs at the same region (9p21.3, ≈22.1 Mb) that are associated with CAD and MI (Helgadottir et al., 2007; McPherson et al., 2007; Wellcome Trust Case Control Consortium, 2007). These SNPs are near CDKN2A and CDKN2B, two tumor suppressor genes.
Genome-wide association studies have also been used in mice to find genes affecting plasma HDL cholesterol levels (Pletcher et al., 2004). The availability of dense SNP maps in more than 50 inbred mouse strains and a better understanding of mouse haplotype structures have greatly improved the feasibility and reliability of such studies.
Genome-wide/large-scale association studies can help find new atherosclerosis-regulating genes in three ways. First, the few genes in a usually very narrow associated region can be confirmed and their functions determined in separate populations. Second, an associated region can be used to pin-point candidate genes in a coincident QTL and thereby facilitate identifying its underlying gene. Third, some QTL regions can be selected for large-scale gene association studies by testing dense SNPs in the region.
Using linkage and association studies to find atherosclerosis genes in mice and humans: expectations, limitations and future directions
Traditional models of atherosclerosis have been very useful in shedding light on this very complex disease, especially on the early stages of atherogenesis. However, we need good models for the clinical outcomes of plaque rupture, which cause thrombotic occlusions and myocardial infarction. Additionally, Apoe- and Ldlr-deficient mutants with genetic backgrounds other than C57BL/6 should be produced: the use of such strains along with newly available bioinformatics tools will likely reveal novel atherosclerosis QTL and facilitate identifying their underlying genes (Wang et al., 2005b).
It is noteworthy that finding an atherosclerosis QTL gene may subsequently reveal a new pathway for atherogenesis. Even if the human ortholog of an atherosclerosis-regulating mouse gene is not a good target, other genes in the new pathways identified from genetics and genomics may be.
According to our current knowledge, two major disturbances affect the progression of atherosclerosis: dyslipidemia and inflammatory response. Identifying their genetic determinants may provide new therapies. The use of mouse models to identify genes regulating plasma lipid levels and atherosclerotic lesions has been reviewed recently (Rollins et al., 2006; Wang et al., 2005b; Wang and Paigen, 2005a; Wang and Paigen, 2005b). Two genes identified in human linkage studies of MI (ALOX5AP and MEF2A), the two in mouse linkage studies of atherosclerosis (Alox5 and Tnfsf4 in mice), and the two genes in human genome-wide association studies of MI (LTA and PSMA6) are all involved in immune and inflammatory response, not directly in lipid metabolism. This may not be a coincidence, because when atherosclerosis or MI is used as an end point, the genes that are historically under selective pressure are easier to find. It is conceivable that genes involved in defense against infection (rather than those involved in influencing plasma lipid and blood pressure levels) have been under greater selective pressure for human beings to reproduce and survive, therefore, their polymorphisms are more tightly associated with inflammatory diseases (such as atherosclerosis) and the associations are easier to find. This observation (that all the six genes found are involved in inflammation) also suggests that genetic determinants of inflammatory pathways account for significant part of population variance of atherosclerotic disease incidence.
Genetic linkage, candidate gene association, and genome-wide associations studies all have their advantages and limitations. Genetic linkage studies have been very successful in finding genes for diseases with simple Mendelian inheritance. However, although there are more and more successfully examples in the last few years (discussed above), finding human atherosclerosis genes from linkage studies remains difficult, and testing the usually 100–200 genes in a QTL remains a daunting task. The fact that only 3 out of the 40 QTL for the same atherosclerosis disease have been replicated, and most of the variants identified from genome-wide association studies are not in any previously found QTL, reflects the nature and complexity of atherosclerotic disease: etiological heterogeneity of the disease, many influencing genes each with very small effect, and strong influence of environmental factors (life-style, diet, social-economical status, etc). Adding to the complexity is the fact that genes affecting a trait tend to be clustered in chromosomes and they may interact with each other to cause a change in the phenotype, making positional cloning difficult. As recently shown in an elegant study by Galzalpour et al., a gene identified by positional cloning may not be the only one that affects the phenotype of the QTL (Ghazalpour et al., 2006). By breaking the original Chr 6 CAST.B6 “congenic” region into 16 subregions, the authors found that, in addition to Alox5, two to four other genes in the original Chr 6 QTL region affect atherosclerosis. This suggests that the original congenic strain is not coisogenic, and the time-consuming process of generating subcongenic lines is actually powerful for finding the real and multiple QTL genes. “Peakwide mapping” (Wang et al., 2007), ie. doing association studies for genes in the QTL peak, as discussed above, should be useful for identifying multiple causal genes for a QTL. Using mouse-human comparative genomics approach should also increase the chance of finding human atherosclerosis genes, as demonstrated in the case of OX40L (Wang et al., 2005c). Candidate gene association studies are more powerful and successful in testing whether the gene of interest confers risk to atherosclerotic disease. However, only genes suspected of affecting atherosclerosis will be tested (therefore this approach is a biased), and significant association from one studies needs to be confirmed in additional studies to exclude false positives, which is often seen in candidate gene association studies. Another limitation of this approach is that the association itself does not tell whether and how the polymorphisms affect the functions of the genes to be tested, such as in the case of ALOX5, ALOX5AP and OX40L. Ultimately, functional studies in vitro and in vivo are needed to find the causal polymorphisms and to evaluate the gene as new therapeutic target. As another unbiased approach, the more recent genome-wide association studies have been very successful in finding unexpected candidate disease genes, including those for atherosclerotic disease. The availability of dense SNP sets will likely increase the power of this approach further. However, at this time the cost of doing this study is still high (millions of USD). More recently, the technological advances, most notably the rapid and low-cost microarray and genotyping methods, have made it possible to treat gene expression data as phenotypes and locate QTL for them; these are so-call expression QTL (eQTL). This allows quantifying the expression of tens of thousands of genes simultaneously in an unbiased way. Combining eQTL and traditional clinical QTL (cQTL; for example atherosclerosis and HDL QTL), candidate genes can be found for further testing. For example, if a cis-eQTL is found within the interval of a cQTL, the cis-eQTL gene would be good candidates for further test (Hubner et al., 2005; Yaguchi et al., 2005). Using this approach, Abcc6 was found as a causal gene for dystrophic cardiac calcification in mice (Meng et al., 2007). We can expect that new atherosclerosis-related genes will be found with this approach in the coming years.
Linkage, candidate gene association, genome-wide association studies and gene expression analysis all have their advantages and limitations. By combining them together, we will have a better chance of defining the complex network underlying atherosclerotic disease.
Translating genome information into medication of atherosclerosis: future perspectives
Researchers are very hopeful that the sequencing of human and model organism genomes, along with the many other technological advances in biomedical research tools, will lead to dramatic improvements in cardiovascular disease therapies. Although two people may have the same MI symptoms, those symptoms may be caused by different combinations of genetic and environmental factors, and may call for different therapies. As such, finding genetic determinants for atherosclerotic disease is crucial for individualized medication. Finding these genes and knowing their functions will greatly facilitate drug target development. Some of the genes and their products may also be used as biomarkers for diagnosis, and for stratifying patients according to genetic risk factors in order to take early preventive measures.
Recently, the DeCode investigators performed a randomized, prospective, placebo-controlled, crossover phase II trial in 191 MI patients with at-risk variants of either the ALOX5AP or the leukotriene A4 hydrolase gene to determine the therapeutic effects of DG-031, an ALOX5AP inhibitor (Hakonarson et al., 2005). In patients with specific at-risk variants of the two genes in the leukotriene pathway, DG-031 effectively reduces leukotriene B4 production by activated neutrophils and increases urinary leukotriene E4 levels. DG-031 also suppresses three biomarkers of increased MI risk (C-reactive protein, intercellular adhesion molecule, myeloperoxidase). This trial was the first to use a gene-specific and individualized approach to treat atherosclerosis, and these preliminary results illustrate the potential to translate genomics information into novel diagnostics and treatments.
Obviously, atherosclerotic disease is very complex, and a large amount of research is required before genetics can be translated into clinical practice (Arnett et al., 2007). The genetic approaches discussed in this review will greatly accelerate the pace with which we acquire that knowledge. Together with clinical data, this knowledge will help us better understand cardiovascular pharmacogenetics and pharmacogenomics, and facilitate the development of individualized diagnosis, preventatives and treatments for atherosclerosis (Arnett et al., 2007).
Acknowledgments
This work was supported by grants from NIH (HL74086, HL77796, HL81162) and from the American Heart Association (AHA 0430381N). The authors thank Jennifer L. Torrance for preparing the figures, and Raymond Lambert for helping to prepare the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiello RJ, Bourassa PA, Lindsey S, Weng W, Freeman A, Showell HJ. Leukotriene B4 receptor antagonism reduces monocytic foam cells in mice. Arterioscler Thromb Vasc Biol. 2002;22:443–449. doi: 10.1161/hq0302.105593. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN. MEF2A sequence variants and coronary artery disease: a change of heart? J Clin Invest. 2005;115:831–833. doi: 10.1172/JCI200524715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett DK, Baird AE, Barkley RA, Basson CT, Boerwinkle E, Ganesh SK, Herrington DM, Hong Y, Jaquish C, McDermott DA, O'Donnell CJ. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: a scientific statement from the American Heart Association Council on Epidemiology and Prevention, the Stroke Council, and the Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation. 2007;115:2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- Bhagavatula MR, Fan C, Shen GQ, Cassano J, Plow EF, Topol EJ, Wang Q. Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet. 2004;13:3181–3188. doi: 10.1093/hmg/ddh329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschger Seidelmann S, De Luca C, Leibel RL, Breslow JL, Tall AR, Welch CL. Quantitative Trait Locus Mapping of Genetic Modifiers of Metabolic Syndrome and Atherosclerosis in Low-Density Lipoprotein Receptor-Deficient Mice. Identification of a Locus for Metabolic Syndrome and Increased Atherosclerosis on Chromosome 4. Arterioscler Thromb Vasc Biol. 2005;25:1–8. doi: 10.1161/01.ATV.0000149146.32385.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, et al. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–214. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
- Brousseau ME. Emerging role of high-density lipoprotein in the prevention of cardiovascular disease. Drug Discov Today. 2005;10:1095–1101. doi: 10.1016/S1359-6446(05)03514-2. [DOI] [PubMed] [Google Scholar]
- Burgess JK, Carlin S, Pack RA, Arndt GM, Au WW, Johnson PR, Black JL, Hunt NH. Detection and characterization of OX40 ligand expression in human airway smooth muscle cells: a possible role in asthma? J Allergy Clin Immunol. 2004;113:683–689. doi: 10.1016/j.jaci.2003.12.311. [DOI] [PubMed] [Google Scholar]
- Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, Xie MH, Gurney A, Bodary S, Liang XH, et al. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. 2002;277:17281–17290. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- Chen AI, McAdam AJ, Buhlmann JE, Scott S, Lupher ML, Jr, Greenfield EA, Baum PR, Fanslow WC, Calderhead DM, Freeman GJ, Sharpe AH. Ox40-ligand has a critical costimulatory role in dendritic cell:T cell interactions. Immunity. 1999;11:689–698. doi: 10.1016/s1074-7613(00)80143-0. [DOI] [PubMed] [Google Scholar]
- Chiodini BD, Lewis CM. Meta-analysis of 4 coronary heart disease genome-wide linkage studies confirms a susceptibility locus on chromosome 3q. Arterioscler Thromb Vasc Biol. 2003;23:1863–1868. doi: 10.1161/01.ATV.0000093281.10213.DB. [DOI] [PubMed] [Google Scholar]
- Cipollone F, Ferri C, Desideri G, Paloscia L, Materazzo G, Mascellanti M, Fazia M, Iezzi A, Cuccurullo C, Pini B, et al. Preprocedural level of soluble CD40L is predictive of enhanced inflammatory response and restenosis after coronary angioplasty. Circulation. 2003;108:2776–2782. doi: 10.1161/01.CIR.0000103700.05109.0D. [DOI] [PubMed] [Google Scholar]
- Clarke R, Xu P, Bennett D, Lewington S, Zondervan K, Parish S, Palmer A, Clark S, Cardon L, Peto R, et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2:e107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinayo VV, Qiao JH, Wang X, Krass KL, Schadt E, Lusis AJ, Drake TA. Genetic loci for diet-induced atherosclerosis lesions and plasma lipids in mice. Mamm Genome. 2003;14:464–471. doi: 10.1007/s00335-002-2187-2. [DOI] [PubMed] [Google Scholar]
- Connelly JJ, Wang T, Cox JE, Haynes C, Wang L, Shah SH, Crosslin DR, Hale AB, Nelson S, Crossman DC, et al. GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet. 2006;2:e139. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansky HM, Shu P, Donavan M, Montagno J, Nagle DL, Smutko JS, Roy N, Whiteing S, Barrios J, McBride TJ, et al. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winther MP, Kanters E, Kraal G, Hofker MH. Nuclear factor kappaB signaling in atherogenesis. Arterioscler Thromb Vasc Biol. 2005;25:904–914. doi: 10.1161/01.ATV.0000160340.72641.87. [DOI] [PubMed] [Google Scholar]
- DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. doi: 10.1016/j.tig.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Dwyer JH, Allayee H, Dwyer KM, Fan J, Wu H, Mar R, Lusis AJ, Mehrabian M. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N Engl J Med. 2004;350:29–37. doi: 10.1056/NEJMoa025079. [DOI] [PubMed] [Google Scholar]
- Farrall M, Green FR, Peden JF, Olsson PG, Clarke R, Hellenius ML, Rust S, Lagercrantz J, Franzosi MG, Schulte H, et al. Genome-wide mapping of susceptibility to coronary artery disease identifies a novel replicated locus on chromosome 17. PLoS Genet. 2006;2:e72. doi: 10.1371/journal.pgen.0020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CS, Cupples LA, Chazaro I, Polak JF, Wolf PA, D'Agostino RB, Ordovas JM, O'Donnell CJ. Genomewide linkage analysis for internal carotid artery intimal medial thickness: evidence for linkage to chromosome 12. Am J Hum Genet. 2004;74:253–261. doi: 10.1086/381559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, et al. A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet. 2001;10:2751–2765. doi: 10.1093/hmg/10.24.2751. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Wang X, Lusis AJ, Mehrabian M. Complex inheritance of the 5-lipoxygenase locus influencing atherosclerosis in mice. Genetics. 2006;173:943–951. doi: 10.1534/genetics.106.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez P, Garcia-Castro M, Reguero JR, Batalla A, Ordonez AG, Palop RL, Lozano I, Montes M, Alvarez V, Coto E. The Pro279Leu variant in the transcription factor MEF2A is associated with myocardial infarction. J Med Genet. 2006;43:167–169. doi: 10.1136/jmg.2005.035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S, Sveinbjornsdottir S, Jonsson HH, Jakobsson F, Einarsdottir E, Agnarsson U, Shkolny D, Einarsson G, Gudjonsdottir HM, Valdimarsson EM, et al. Localization of a susceptibility gene for common forms of stroke to 5q12. Am J Hum Genet. 2002;70:593–603. doi: 10.1086/339252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson G, Matthiasson SE, Arason H, Johannsson H, Runarsson F, Bjarnason H, Helgadottir K, Thorisdottir S, Ingadottir G, Lindpaintner K, et al. Localization of a gene for peripheral arterial occlusive disease to chromosome 1p31. Am J Hum Genet. 2002;70:586–592. doi: 10.1086/339251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H, Thorvaldsson S, Helgadottir A, Gudbjartsson D, Zink F, Andresdottir M, Manolescu A, Arnar DO, Andersen K, Sigurdsson A, et al. Effects of a 5-lipoxygenase-activating protein inhibitor on biomarkers associated with risk of myocardial infarction: a randomized trial. Jama. 2005;293:2245–2256. doi: 10.1001/jama.293.18.2245. [DOI] [PubMed] [Google Scholar]
- Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- Harrap SB, Zammit KS, Wong ZY, Williams FM, Bahlo M, Tonkin AM, Anderson ST. Genome-wide linkage analysis of the acute coronary syndrome suggests a locus on chromosome 2. Arterioscler Thromb Vasc Biol. 2002;22:874–878. doi: 10.1161/01.atv.0000016258.40568.f1. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, McAdam B, Winkelmann BR, Wiseman AH, Muhlestein JB, et al. A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet. 2004;75:436–447. doi: 10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Gretarsdottir S, St Clair D, Manolescu A, Cheung J, Thorleifsson G, Pasdar A, Grant SF, Whalley LJ, Hakonarson H, et al. Association between the gene encoding 5-lipoxygenase-activating protein and stroke replicated in a Scottish population. Am J Hum Genet. 2005;76:505–509. doi: 10.1086/428066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Heller EA, Liu E, Tager AM, Sinha S, Roberts JD, Koehn SL, Libby P, Aikawa ER, Chen JQ, Huang P, et al. Inhibition of atherogenesis in BLT1-deficient mice reveals a role for LTB4 and BLT1 in smooth muscle cell recruitment. Circulation. 2005;112:578–586. doi: 10.1161/CIRCULATIONAHA.105.545616. [DOI] [PubMed] [Google Scholar]
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- Ishimori N, Li R, Kelmenson PM, Korstanje R, Walsh KA, Churchill GA, Forsman-Semb K, Paigen B. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6J and 129S1/SvImJ. Arterioscler Thromb Vasc Biol. 2004;24:161–166. doi: 10.1161/01.ATV.0000104027.52895.D7. [DOI] [PubMed] [Google Scholar]
- Jawien J, Gajda M, Rudling M, Mateuszuk L, Olszanecki R, Guzik TJ, Cichocki T, Chlopicki S, Korbut R. Inhibition of five lipoxygenase activating protein (FLAP) by MK-886 decreases atherosclerosis in apoE/LDLR-double knockout mice. Eur J Clin Invest. 2006;36:141–146. doi: 10.1111/j.1365-2362.2006.01606.x. [DOI] [PubMed] [Google Scholar]
- Kajimoto K, Shioji K, Ishida C, Iwanaga Y, Kokubo Y, Tomoike H, Miyazaki S, Nonogi H, Goto Y, Iwai N. Validation of the association between the gene encoding 5-lipoxygenase-activating protein and myocardial infarction in a Japanese population. Circ J. 2005a;69:1029–1034. doi: 10.1253/circj.69.1029. [DOI] [PubMed] [Google Scholar]
- Kajimoto K, Shioji K, Tago N, Tomoike H, Nonogi H, Goto Y, Iwai N. Assessment of MEF2A mutations in myocardial infarction in Japanese patients. Circ J. 2005b;69:1192–1195. doi: 10.1253/circj.69.1192. [DOI] [PubMed] [Google Scholar]
- Kaushal R, Pal P, Alwell K, Haverbusch M, Flaherty M, Moomaw C, Sekar P, Kissela B, Kleindorfer D, Chakraborty R, et al. Hum Genet. 2007. Association of ALOX5AP with ischemic stroke: a population-based case-control study. [DOI] [PubMed] [Google Scholar]
- Koch W, Hoppmann P, Mueller JC, Schomig A, Kastrati A. No association of polymorphisms in the gene encoding 5-lipoxygenase-activating protein and myocardial infarction in a large central European population. Genet Med. 2007;9:123–129. doi: 10.1097/gim.0b013e318030c9c5. [DOI] [PubMed] [Google Scholar]
- Kopf M, Ruedl C, Schmitz N, Gallimore A, Lefrang K, Ecabert B, Odermatt B, Bachmann MF. OX40-deficient mice are defective in Th cell proliferation but are competent in generating B cell and CTL Responses after virus infection. Immunity. 1999;11:699–708. doi: 10.1016/s1074-7613(00)80144-2. [DOI] [PubMed] [Google Scholar]
- Korstanje R, Eriksson P, Samnegard A, Olsson PG, Forsman-Semb K, Sen S, Churchill GA, Rollins J, Harris S, Hamsten A, Paigen B. Locating Ath8, a locus for murine atherosclerosis susceptibility and testing several of its candidate genes in mice and humans. Atherosclerosis. 2004;177:443–450. doi: 10.1016/j.atherosclerosis.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Koster A, Chao YB, Mosior M, Ford A, Gonzalez-DeWhitt PA, Hale JE, Li D, Qiu Y, Fraser CC, Yang DD, et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- Kotani A, Hori T, Matsumura Y, Uchiyama T. Signaling of gp34 (OX40 ligand) induces vascular endothelial cells to produce a CC chemokine RANTES/CCL5. Immunol Lett. 2002;84:1–7. doi: 10.1016/s0165-2478(02)00082-2. [DOI] [PubMed] [Google Scholar]
- Lange LA, Lange EM, Bielak LF, Langefeld CD, Kardia SL, Royston P, Turner ST, Sheedy PF, 2nd, Boerwinkle E, Peyser PA. Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler Thromb Vasc Biol. 2002;22:418–423. doi: 10.1161/hq0302.105721. [DOI] [PubMed] [Google Scholar]
- Laxton R, Pearce E, Kyriakou T, Ye S. Association of the lymphotoxin-alpha gene Thr26Asn polymorphism with severity of coronary atherosclerosis. Genes Immun. 2005;6:539–541. doi: 10.1038/sj.gene.6364236. [DOI] [PubMed] [Google Scholar]
- Lohmussaar E, Gschwendtner A, Mueller JC, Org T, Wichmann E, Hamann G, Meitinger T, Dichgans M. ALOX5AP gene and the PDE4D gene in a central European population of stroke patients. Stroke. 2005;36:731–736. doi: 10.1161/01.STR.0000157587.59821.87. [DOI] [PubMed] [Google Scholar]
- Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- Mangino M, Braund P, Singh R, Steeds R, Thompson JR, Channer K, Samani NJ. LGALS2 functional variant rs7291467 is not associated with susceptibility to myocardial infarction in Caucasians. Atherosclerosis. 2006 doi: 10.1016/j.atherosclerosis.2006.10.004. in press. [DOI] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M, Allayee H, Wong J, Shi W, Wang XP, Shaposhnik Z, Funk CD, Lusis AJ, Shih W. Identification of 5-lipoxygenase as a major gene contributing to atherosclerosis susceptibility in mice. Circ Res. 2002;91:120–126. doi: 10.1161/01.res.0000028008.99774.7f. [DOI] [PubMed] [Google Scholar]
- Mehrabian M, Wong J, Wang X, Jiang Z, Shi W, Fogelman AM, Lusis AJ. Genetic locus in mice that blocks development of atherosclerosis despite extreme hyperlipidemia. Circ Res. 2001;89:125–130. doi: 10.1161/hh1401.093458. [DOI] [PubMed] [Google Scholar]
- Meng H, Vera I, Che N, Wang X, Wang SS, Ingram-Drake L, Schadt EE, Drake TA, Lusis AJ. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc Natl Acad Sci U S A. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschia JF, Brott TG, Brown RD, Jr, Crook R, Worrall BB, Kissela B, Brown WM, Rich SS, Case LD, Evans EW, et al. Phosphodiesterase 4D and 5-lipoxygenase activating protein in ischemic stroke. Ann Neurol. 2005;58:351–361. doi: 10.1002/ana.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Sato H, Sakata Y, Ohnishi Y, Hishida E, Kinjo K, Nakatani D, Shimizu M, Kondo H, Tanaka T, et al. Impact of atherosclerosis-related gene polymorphisms on mortality and recurrent events after myocardial infarction. Atherosclerosis. 2006;185:400–405. doi: 10.1016/j.atherosclerosis.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Monaco C, Paleolog E. Nuclear factor kappaB: a potential therapeutic target in atherosclerosis and thrombosis. Cardiovasc Res. 2004;61:671–682. doi: 10.1016/j.cardiores.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Mu JL, Naggert JK, Svenson KL, Collin GB, Kim JH, McFarland C, Nishina PM, Levine DM, Williams KJ, Paigen B. Quantitative trait loci analysis for the differences in susceptibility to atherosclerosis and diabetes between inbred mouse strains C57BL/6J and C57BLKS/J. J Lipid Res. 1999;40:1328–1335. [PubMed] [Google Scholar]
- Murata K, Ishii N, Takano H, Miura S, Ndhlovu LC, Nose M, Noda T, Sugamura K. Impairment of antigen-presenting cell function in mice lacking expression of OX40 ligand. J Exp Med. 2000;191:365–374. doi: 10.1084/jem.191.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson-Ardnor S, Wiklund PG, Lindgren P, Nilsson AK, Janunger T, Escher SA, Hallbeck B, Stegmayr B, Asplund K, Holmberg D. Linkage of ischemic stroke to the PDE4D region on 5q in a Swedish population. Stroke. 2005;36:1666–1671. doi: 10.1161/01.STR.0000174188.04716.8d. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Inoue K, Sato H, Iida A, Ohnishi Y, Sekine A, Sato H, Odashiro K, Nobuyoshi M, Hori M, et al. Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-alpha secretion in vitro. Nature. 2004;429:72–75. doi: 10.1038/nature02502. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- Paigen B. Genetics of responsiveness to high-fat and high-cholesterol diets in the mouse. Am J Clin Nutr. 1995;62:458S–462S. doi: 10.1093/ajcn/62.2.458S. [DOI] [PubMed] [Google Scholar]
- Paigen B, Albee D, Holmes PA, Mitchell D. Genetic analysis of murine strains C57BL/6J and C3H/HeJ to confirm the map position of Ath-1, a gene determining atherosclerosis susceptibility. Biochem Genet. 1987a;25:501–511. doi: 10.1007/BF00554352. [DOI] [PubMed] [Google Scholar]
- Paigen B, Mitchell D, Holmes PA, Albee D. Genetic analysis of strains C57BL/6J and BALB/cJ for Ath-1, a gene determining atherosclerosis susceptibility in mice. Biochem Genet. 1987b;25:881–892. doi: 10.1007/BF00502607. [DOI] [PubMed] [Google Scholar]
- Paigen B, Mitchell D, Reue K, Morrow A, Lusis AJ, LeBoeuf RC. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc Natl Acad Sci USA. 1987c;84:3763–3767. doi: 10.1073/pnas.84.11.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, Terwilliger JD, Kempas E, Daly M, Lilja H, et al. Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet. 2000;67:1481–1493. doi: 10.1086/316902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan SA, Beier DR, Higgins DC, Paigen B. Confirmation and high resolution mapping of an atherosclerosis susceptibility gene in mice on Chromosome 1. Mamm Genome. 2002;13:548–553. doi: 10.1007/s00335-002-2196-1. [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROCARDIS-Consortium. A trio family study showing association of the lymphotoxin-alpha N26 (804A) allele with coronary artery disease. Eur J Hum Genet. 2004;12:770–774. doi: 10.1038/sj.ejhg.5201244. [DOI] [PubMed] [Google Scholar]
- Qiu H, Gabrielsen A, Agardh HE, Wan M, Wetterholm A, Wong CH, Hedin U, Swedenborg J, Hansson GK, Samuelsson B, et al. Expression of 5-lipoxygenase and leukotriene A4 hydrolase in human atherosclerotic lesions correlates with symptoms of plaque instability. Proc Natl Acad Sci U S A. 2006;103:8161–8166. doi: 10.1073/pnas.0602414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ria M, Eriksson P, Boquist S, Ericsson CG, Hamsten A, Lagercrantz J. Human genetic evidence that OX40 is implicated in myocardial infarction. Biochem Biophys Res Commun. 2006;339:1001–1006. doi: 10.1016/j.bbrc.2005.11.092. [DOI] [PubMed] [Google Scholar]
- Rollins J, Chen Y, Paigen B, Wang X. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc Med. 2006;16:220–234. doi: 10.1016/j.tcm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Samani NJ, Burton P, Mangino M, Ball SG, Balmforth AJ, Barrett J, Bishop T, Hall A. A genomewide linkage study of 1,933 families affected by premature coronary artery disease: The British Heart Foundation (BHF) Family Heart Study. Am J Hum Genet. 2005;77:1011–1020. doi: 10.1086/498653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D, Ellis SG, Rowland CM, Malloy MJ, Luke MM, Iakoubova OA, Pullinger CR, Cassano J, Aouizerat BE, Fenwick RG, et al. Identification of four gene variants associated with myocardial infarction. Am J Hum Genet. 2005;77:596–605. doi: 10.1086/491674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D, Rowland CM, Louie JZ, Luke MM, Bare LA, Bolonick JI, Young BA, Catanese JJ, Stiggins CF, Pullinger CR, et al. Gene variants of VAMP8 and HNRPUL1 are associated with early-onset myocardial infarction. Arterioscler Thromb Vasc Biol. 2006;26:1613–1618. doi: 10.1161/01.ATV.0000226543.77214.e4. [DOI] [PubMed] [Google Scholar]
- Smith JD, Bhasin JM, Baglione J, Settle M, Xu Y, Barnard J. Atherosclerosis susceptibility loci identified from a strain intercross of apolipoprotein E-deficient mice via a high-density genome scan. Arterioscler Thromb Vasc Biol. 2006;26:597–603. doi: 10.1161/01.ATV.0000201044.33220.5c. [DOI] [PubMed] [Google Scholar]
- Stewart-Phillips JL, Lough J, Skamene E. ATH-3, a new gene for atherosclerosis in the mouse. Clin Invest Med. 1989;12:121–126. [PubMed] [Google Scholar]
- Su Z, Li Y, James JC, McDuffie M, Matsumoto AH, Helm GA, Weber JL, Lusis AJ, Shi W. Quantitative trait locus analysis of atherosclerosis in an intercross between C57BL/6 and C3H mice carrying the mutant apolipoprotein E gene. Genetics. 2006;172:1799–1807. doi: 10.1534/genetics.105.051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, Jala VR, Mathis S, Suttles J, Zacharias W, Ahamed J, Ali H, Tseng MT, Haribabu B. Role of leukotriene B4 receptors in the development of atherosclerosis: potential mechanisms. Arterioscler Thromb Vasc Biol. 2004;24:369–375. doi: 10.1161/01.ATV.0000110503.16605.15. [DOI] [PubMed] [Google Scholar]
- Sugamura K, Ishii N, Weinberg AD. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat Rev Immunol. 2004;4:420–431. doi: 10.1038/nri1371. [DOI] [PubMed] [Google Scholar]
- Teupser D, Breslow JL. Quantitative trait locus mapping of atherosclerotic lesions in a cross between B6.129-Ldlr−/− and FVB.129-Ldlr−/− mice. Paper presented at the 5th Annual Conference on Arteriosclerosis, Thrombosis and Vascular Biology; San Francisco. May 6–8, 2004.2004. [Google Scholar]
- Teupser D, Tan M, Persky AD, Breslow JL. Atherosclerosis quantitative trait loci are sex- and lineage-dependent in an intercross of C57BL/6 and FVB/N low-density lipoprotein receptor−/− mice. Proc Natl Acad Sci U S A. 2006;103:123–128. doi: 10.1073/pnas.0509570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Farmer JA. Optimal low-density lipoprotein levels: evidence from epidemiology and clinical trials. Curr Atheroscler Rep. 2006;8:157–162. doi: 10.1007/s11883-006-0053-5. [DOI] [PubMed] [Google Scholar]
- Turner ST, Peyser PA, Kardia SL, Bielak LF, Sheedy PF, 3rd, Boerwinkle E, de Andrade M. Genomic loci with pleiotropic effects on coronary artery calcification. Atherosclerosis. 2005;185:340–346. doi: 10.1016/j.atherosclerosis.2005.06.010. [DOI] [PubMed] [Google Scholar]
- van Wanrooij EJ, van Puijvelde GH, de Vos P, Yagita H, van Berkel TJ, Kuiper J. Interruption of the Tnfrsf4/Tnfsf4 (OX40/OX40L) pathway attenuates atherogenesis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:204–210. doi: 10.1161/01.ATV.0000251007.07648.81. [DOI] [PubMed] [Google Scholar]
- Veillard NR, Kwak B, Pelli G, Mulhaupt F, James RW, Proudfoot AE, Mach F. Antagonism of RANTES receptors reduces atherosclerotic plaque formation in mice. Circ Res. 2004;94:253–261. doi: 10.1161/01.RES.0000109793.17591.4E. [DOI] [PubMed] [Google Scholar]
- Wang D, Yang H, Quinones MJ, Bulnes-Enriquez I, Jimenez X, De La Rosa R, Modilevsky T, Yu K, Li Y, Taylor KD, et al. A genome-wide scan for carotid artery intima-media thickness: the Mexican-American Coronary Artery Disease family study. Stroke. 2005a;36:540–545. doi: 10.1161/01.STR.0000155746.65185.4e. [DOI] [PubMed] [Google Scholar]
- Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003a;302:1578–1581. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, Harris M, Nelson S, Hale AB, Granger CB, et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Rao S, Shen GQ, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, Topol EJ. Premature myocardial infarction novel susceptibility locus on chromosome 1p34-36 identified by genomewide linkage analysis. Am J Hum Genet. 2004;74:262–271. doi: 10.1086/381560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Shi W, Wang X, Lusis AJ. Identification of a locus in mice tht influences aortic aneurysm formation. Circ Res. 2003b;108:IV–177. [Google Scholar]
- Wang X. T-cell co-stimulators as anti-inflammatory targets for atherosclerotic disease. Future Cardiol. 2006;2:187–195. doi: 10.2217/14796678.2.2.187. [DOI] [PubMed] [Google Scholar]
- Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005b;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paigen B. Genetics of variation in HDL cholesterol in humans and mice. Circ Res. 2005a;96:27–42. doi: 10.1161/01.RES.0000151332.39871.13. [DOI] [PubMed] [Google Scholar]
- Wang X, Paigen B. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr Opin Lipidol. 2005b;16:127–137. doi: 10.1097/01.mol.0000162317.09054.9d. [DOI] [PubMed] [Google Scholar]
- Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, Petros C, Rollins J, Bennet AM, Wiman B, et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet. 2005c;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, Liang CP, Barlow C, Dansky H, Breslow JL, Tall AR. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc Natl Acad Sci USA. 2001;98:7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, Kavaslar N, Ustaszewska A, Doelle H, Schackwitz W, Hebert S, Cohen JC, McPherson R, Pennacchio LA. Lack of MEF2A mutations in coronary artery disease. J Clin Invest. 2005;115:1016–1020. doi: 10.1172/JCI24186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi H, Togawa K, Moritani M, Itakura M. Identification of candidate genes in the type 2 diabetes modifier locus using expression QTL. Genomics. 2005;85:591–599. doi: 10.1016/j.ygeno.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Zee RY, Cheng S, Hegener HH, Erlich HA, Ridker PM. Genetic variants of arachidonate 5-lipoxygenase-activating protein, and risk of incident myocardial infarction and ischemic stroke: a nested case-control approach. Stroke. 2006;37:2007–2011. doi: 10.1161/01.STR.0000229905.25080.01. [DOI] [PubMed] [Google Scholar]
- Zhao L, Moos MP, Grabner R, Pedrono F, Fan J, Kaiser B, John N, Schmidt S, Spanbroek R, Lotzer K, et al. The 5-lipoxygenase pathway promotes pathogenesis of hyperlipidemia-dependent aortic aneurysm. Nat Med. 2004;10:966–973. doi: 10.1038/nm1099. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Kitsios G. Identification of chromosomal regions linked to premature myocardial infarction: a meta-analysis of whole-genome searches. J Hum Genet. 2006;51:1015–1021. doi: 10.1007/s10038-006-0053-x. [DOI] [PubMed] [Google Scholar]