Summary
The mammalian Sirtuin proteins contain seven family members that are homologous to yeast Sir2. Here we show that Sirt2, a cytoplasmic sirtuin, is the most abundant sirtuin in adipocytes, its expression is down regulated during preadipocyte differentiation in 3T3-L1 cells. Over-expression of Sirt2 inhibits differentiation, whereas reducing Sirt2 expression promotes adipogenesis. Both effects are accompanied by corresponding changes in the expression of PPARγ, C/EBPα and genes marking terminal adipocyte differentiation, such as Glut4, aP2, and fatty acid synthase. At the molecular level, reducing Sirt2 in 3T3-L1 adipocytes acts by promoting acetylation of FoxO1. This occurs as the result of direct interaction between Sirt2 and FoxO1, and enhances insulin-stimulated phosphorylation of FoxO1, which in turn regulates FoxO1 nuclear and cytosolic localization. Thus, Sirt2 acts as an important regulator of adipocyte differentiation through control of FoxO1 acetylation/phosphorylation and activity and may contribute to control adipose tissue mass and function.
Introduction
The Sir2 (silent information regulator 2) proteins belong to the family of class III NAD-dependent deacetylases that catalyze a reaction in which NAD and an acetylated substrate are converted into a deacetylated protein, nicotinamide and a novel metabolite O-acetyl ADP-ribose (Tanner et al., 2000). The founding member of the family, Sir2 was originally discovered in yeast as a factor that silences the mating type locus (Imai et al., 2000; Tanny et al., 1999). Sir2 is also involved in telomere regulation, maintenance of genomic integrity and lifespan extension in yeast and similar effects have been shown for its orthologue in C. elegans (Imai et al., 2000; Wang and Tissenbaum, 2006).
In mammals, the homologues of Sir2 have been named sirtuins (Sirt), with seven members in a family termed Sirt1 through Sirt7. They share a conserved central deacetylase domain, but have different N- and C- termini and display distinct subcellular localization suggesting different biological functions (North and Verdin, 2004). Mammalian Sirt1 is most homologous to yeast Sir2 and is found predominantly in the nucleus, consistent with its roles in formation of heterochromatin and gene silencing by histone deacetylation. In mammalian cells, instead of genome silencing, Sirt1 often promotes gene transcription by deacetylating specific transcription factors, corepressors, and coactivators, including p53, PGC-1α, NF-kB, MyoD and members of the FoxO family (Daitoku et al., 2004; Fulco et al., 2003; Luo et al., 2001; Nemoto et al., 2005; Yeung et al., 2004). In adipocytes, Sirt1 acts as an inhibitor of adipogenesis by interacting with PPARγ co-repressor NcoR and SMART thereby repressing PPARγ activity (Picard et al., 2004).
Much less is known about the function of the other sirtuins. In contrast to Sirt1, mammalian Sirt2 is localized mainly in the cytoplasm. Studies in mammalian cells have suggested that Sirt2 may play a role in cell cycle regulation and be involved in cytoskeleton organization by targeting the cytoskeletal protein tubulin (North et al., 2003). The yeast orthologue of Sirt2, Hst2 has been shown to extend lifespan by a mechanism independent of Sir2/Hst1 (Lamming et al., 2005). Functional studies showed that Sirt3 deacetylates acetyl-CoA synthase 2 (ACS2) and regulates its activity (Hallows et al., 2006; Schwer et al., 2006). Sirt3 also appears to be involved in longevity (Rose et al., 2003). The functions of Sirt 4-7 are less clear; recent reports have shown that Sirt6 may also be involved in aging in mice, while Sirt7 appear to regulate DNA pol I transcription (Ford et al., 2006; Mostoslavsky et al., 2006).
Mammalian forkhead transcription factors of class O (FoxO) include: FoxO1, FoxO3a, and FoxO4, and have been shown to be involved with wide range of cellular processes, such as DNA repair, cell cycle control, stress resistance, apoptosis, and metabolism (Barthel et al., 2005; Furukawa-Hibi et al., 2005). Among all FoxO members, FoxO1 appears to have an important role in adipocyte differentiation acting as an inhibitor of adipogenesis at an early phase of the differentiation process (Nakae et al., 2003). In this context, the enzyme phosphatidylinositol 3-kinase (PI-kinase), which is stimulated by insulin and certain cytokines and growth factors, can negatively regulate FoxOs (Zhang et al., 2002). This inhibitory effect of insulin is mainly mediated by Akt/PKB phosphorylation of FoxO, which promotes the trafficking of FoxO from the nucleus to the cytosol. The transcriptional activity of FoxO proteins can also be regulated by acetylation and deacetylation. FoxO1 can be acetylated by CBP acetyl-transferase, and Sirt1 has been shown to deacetylate FoxO1 and regulate its activity, especially under conditions of stress (Matsuzaki et al., 2005; van der Heide and Smidt, 2005). Recently, it has been shown that the extent of deacetylation of FoxO1 can affect its phosphorylation and DNA binding activity to target gene promoters (Matsuzaki et al., 2005).
In the present study, we demonstrate that Sirt2 mRNA is the more abundant than that of other sirtuins in adipocytes in vivo and in culture and that Sirt2 exerts an inhibitory effect on adipocyte differentiation. This is mediated by regulating FoxO1 deacetylation, leading to changes in FoxO1 phosphorylation, increased nuclear localization and inhibition of adipogenesis. Thus, activators of Sirt2 could provide novel therapeutics of obesity and its complications.
Results
Sirt Isoforms Expression During Adipocyte Differentiation of 3T3-L1 Cells
Different isoforms of mammalian Sirt proteins are expressed in adipose tissue and 3T3-L1 preadipocytes and exhibit different patterns of change during differentiation. Affymetrix microarrays performed on isolated adipocyte indicated that Sirt1, Sirt2 and Sirt3 were all expressed in adipocytes and that the level of Sirt2 was much higher than that of Sirt1 or Sirt3 (Figure1A, top panel). Using quantitative realtime PCR with cDNA standard curves for each isoform as described in Methods, the molar amounts of different Sirt transcripts per microgram of total RNA were obtained. As shown in Figure 1A lower panel, the molar amount of Sirt2 mRNA per microgram total RNA in 3T3-L1 preadipocytes was 4-5 times that of Sirt1 and 6-7 times of Sirt3. During the first 2 days of differentiation, i.e. the induction phase, levels of both Sirt1 and Sirt2 mRNA decreased by 60-70% and then remained stable for the remainder of the time course of differentiation (Figure 1B, top and middle panels). Sirt3 mRNA on the other hand, started at a low level compared to both Sirt2 and Sirt1, then increased by 3-4 fold during adipocyte differentiation (Figure 1B, bottom panel).
Figure 1.
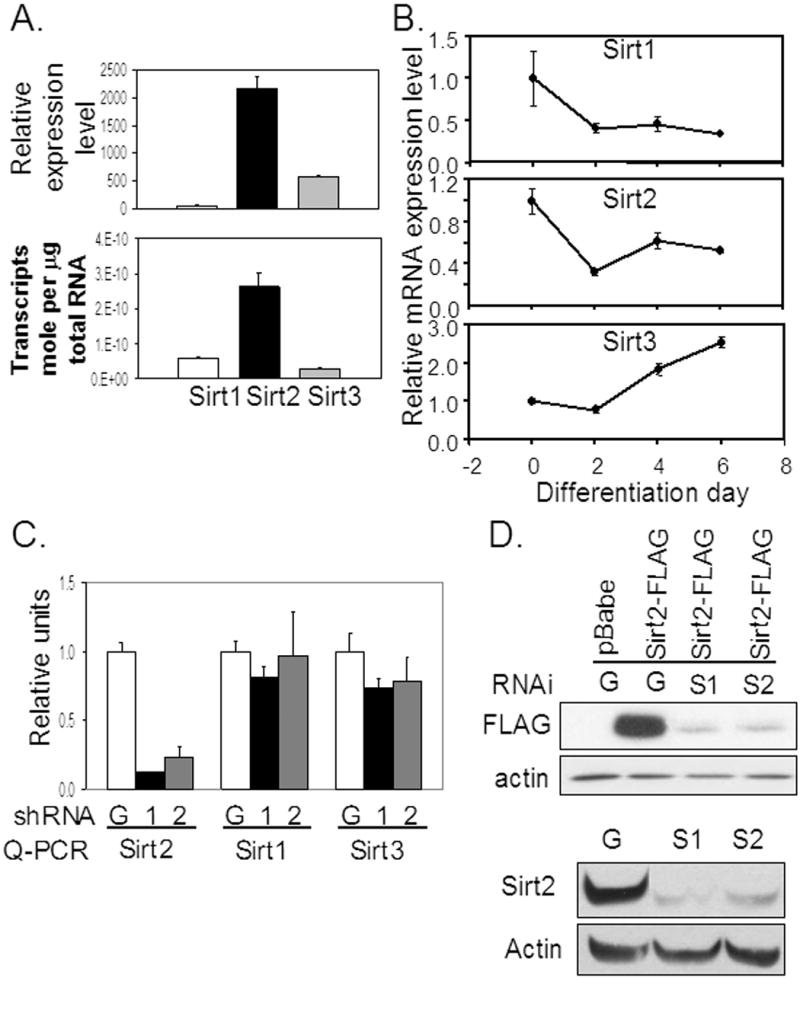
Expression of Sirt2 and stable Sirt2 knockdown in 3T3-L1 preadipocytes. (A) Affymetrix microarray analysis was performed using mRNA isolated from epididymal adipocytes as described previously (Gesta et al., 2006). To confirm these finding, quantitative realtime PCR was performed as described in Material and Methods. (B) Using Realtime PCR, changes in expression of the different Sirt mRNA was assessed during 3T3-L1 white adipocyte differentiation. Sirt2 mRNA was the most abundant in adipocytes and both Sirt1 and Sirt2 had similar pattern of diminishing expression during adipocyte differentiation. (C) shRNA overexpression constructs were generated with pSuper-Retro vector. Two shRNA constructs (S-1 and S-2) were tested targeting different exons of Sirt2 genomic sequence. An shGFP RNAi sequence was used as control. After retroviral infection and selection, 3T3-L1 preadipocytes carrying either shGFP or shSirt2 overexpression constructs were grown to confluence then RNA was extracted to synthesize cDNA and realtime PCR was performed for Sirt 1-3. (D) Transient transfection experiments were done using two different shRNA and control shRNA constructs along with either control pBabe or Sirt2-FLAG overexpression. Both shSirt2 (S1 and S2) effectively knockdown the overexpression of Sirt2-FLAG protein. Endogenous Sirt2 knockdown was also detected by western blot.
Effects of Sirt2 Knockdown and Overexpression in 3T3-L1 Adipocytes
To investigate the potential role of Sirt2 in preadipocytes, we used retroviruses to generate 3T3-L1 stable cell lines carrying either shRNAs targeting endogenous Sirt2 or GFP as a control. Since there was no good antibody available for mouse Sirt2 at the time these experiments were preformed, we used realtime PCR to assess Sirt2 mRNA levels. This revealed that, compared with shGFP cells, cells stably expressing the two shSirt2 retroviruses exhibited an 80-90% knockdown of Sirt2 mRNA, with no significant change in the level of Sirt1 or Sirt3 mRNAs (Figure 1C). Furthermore, when the same two retroviral constructs were transiently co-transfected into HEK293 with a CMV-driven Sirt2-FLAG construct, there was a parallel 80-90% reduction of the tagged Sirt2 protein when compared with cells co-transfected with shGFP (Figure 1D). Thus, the expression of both shSirt2 constructs produced major reductions of Sirt2 at the RNA and protein levels, and this reduction was specific to the Sirt2 isoform. Later commercial availability of Sirt2 antibody allowed us to detect a similar decrease of endogenous Sirt2 protein in shSirt2 cells (Figure 1D)
Pre-adipocytes stably transfected with either shSirt2 or shGFP were then subjected to adipogenic differentiation protocol, and samples from different time points were collected for either RNA or protein analysis. Oil Red O staining during the time course of differentiation confirmed the increased rate and extent of differentiation with increased staining of cells by day 4 indicating more rapid accumulation of lipid in Sirt2 knockdown cells (Figure 2A). As noted above, in control shGFP-expressing cells Sirt2 mRNA expression decreased during the time course of differentiation, while in the shSirt2 expressing cells, endogenous Sirt2 mRNA as assessed by realtime PCR was reduced by 75-80%, and this persisted throughout the time course of adipocyte differentiation (Figure 2B). As expected, Sirt2 knockdown had no significant effect on levels of Sirt1 mRNA or on the change in Sirt1 that occurred during differentiation, consistent with the specificity of Sirt2 knockdown (Figure 2B). By contrast, in the Sirt2 knockdown cells, two transcription factors central to adipogenic differentiation, C/EBPα and PPARγ, both demonstrated significantly accelerated and exaggerated increase in mRNA expression. Thus, C/EBPα mRNA level was elevated more than 3-fold in shSirt2 cells on day 2 after induction compared with control cells (P=0.001), and this difference remained throughout the time course of differentiation (Figure 2B). PPARγ mRNA levels in shSirt2 cells were also 2- to 3- fold higher than in shGFP cells after induction and throughout time course with the greatest increase on day 2 (Figure 2B). Corresponding to elevated early adipogenic transcription factor expression, mRNA levels of various late adipocyte differentiation markers that are downstream C/EBPα and PPARγ (Lane et al., 1999; Qi et al., 2000) were also significantly enhanced in shSirt2 cells during the time course of differentiation. For example, on day 2 after induction, shSirt2 cells had ~ 3 fold higher levels of aP2 mRNA (P=0.002), and ~2 fold higher levels of fatty acid synthase (FAS) (P=0.0014) and Glut 4 mRNA (P=0.0055) comparing to control cells (Figure 2B). On the other hand, expression of FoxO1, showed no significant change at mRNA level at any time point during differentiation (Figure 2B).
Figure 2.
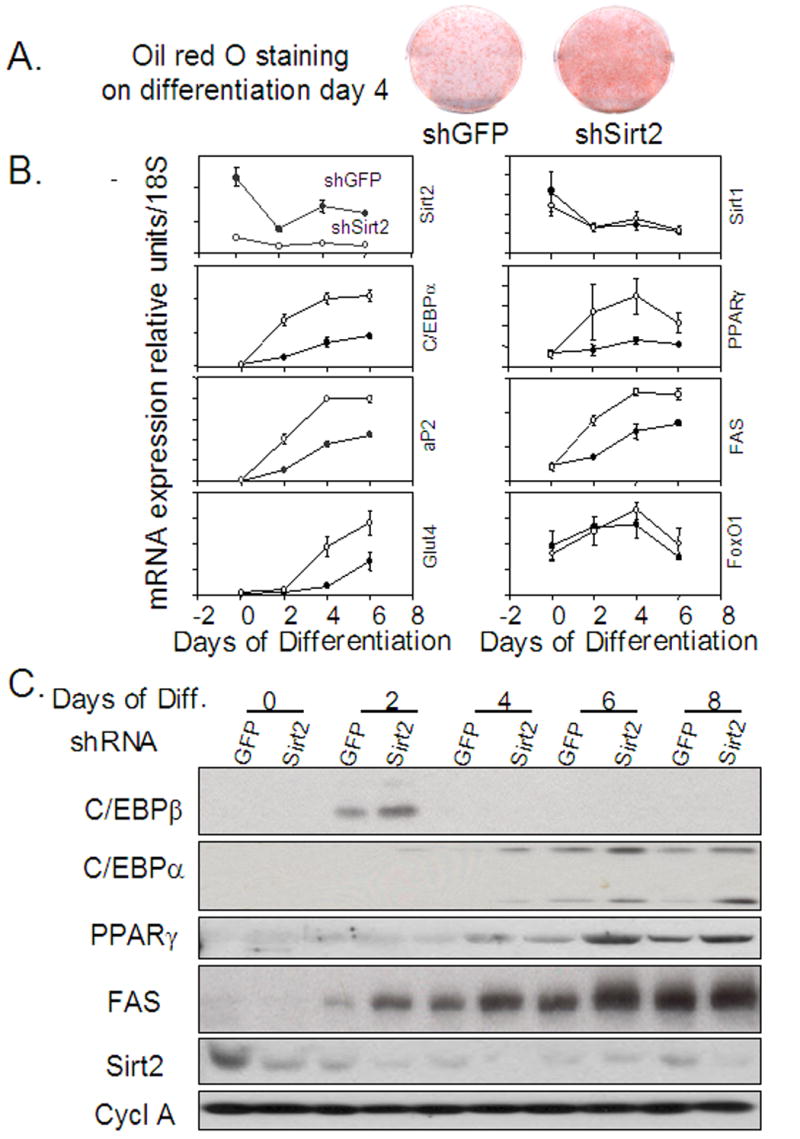
Sirt2 knockdown promotes 3T3L1 adipocyte differentiation. Stable shRNA transfected 3T3-L1 preadipocytes were subjected to differentiation using the standard protocol. Oil Red O staining of shGFP and shSirt2 cells on day 4 of differentiation indicated that shSirt2 had accelerated differentiation with enhanced lipid staining. During 3T3-L1 adipocyte differentiation, shSirt2 cells (empty circles) had consistently lower endogenous Sirt2 mRNA expression compared with shGFP cells (solid circles). The mRNA expression for various differentiation markers was also determined by realtime PCR. The protein expression of different adipocyte differentiation markers was determined by Western blotting.
Western blot analysis of proteins confirmed these effects of Sirt2 knockdown on expression of adipocyte differentiation markers (Figure 2C). On day 2 after induction, there was a 2-fold increase in C/EBPβ and a 5-fold increase in C/EBPα protein in shSirt2 cells compared to control, and this increase in C/EBPα persisted through differentiation, even as levels in the control cells increased. A similar pattern of increased protein expression was observed for PPARγ protein in shSirt2 cells. The increase was even more marked for the late adipocyte differentiation marker, FAS, which was 4-fold elevated at the protein level in shSirt2 cells on day 2 compared to controls, although this difference diminished on day 8 as the cells became mature and FAS expression increased in the control cells (Figure 2C). Endogenous Sirt2 protein expression was consistent with its mRNA expression during differentiation in shGFP cells, while endogenous Sirt2 protein was knocked down in shSirt2 cells.
Opposite effects were observed in 3T3-L1 cells overexpressing Sirt2. Over-expression of Sirt2-FLAG in 3T3-L1 cells inhibited adipocyte differentiation and lipid accumulation compared with empty vector control cells (Figure 3A). Western blot analysis of adipocyte markers, such as PPARγ and FAS, also revealed decreased levels in Sirt2 overexpressing cell line (Figure 3A). As insulin signaling pathway is one of the major pathways that controls adipogenesis and adipocyte differentiation, we tested if the effect of Sirt2 on 3T3-L1 differentiation was due to altered insulin signaling. Acute (10 minutes) insulin stimulation of both control and Sirt2 overexpressing cell lines produced equal phosphorylation responses for Akt, p42/p44 MAP kinase and p38 MAP kinase (Figure 3B). Thus, overexpression of Sirt2 in 3T3-L1 cells inhibits the normal adipogenic process, and this effect occurs without a change in upstream insulin signaling. Conversely, reducing Sirt2 expression enhanced the program of adipogenic gene expressions at the mRNA and protein levels, and this is associated with enhanced lipid accumulation. The subcellular localization of Sirt2-FLAG overexpression is similar to previous reports that Sirt2 is mainly a cytoplasmic protein (Supplemental figure1)
Figure 3.
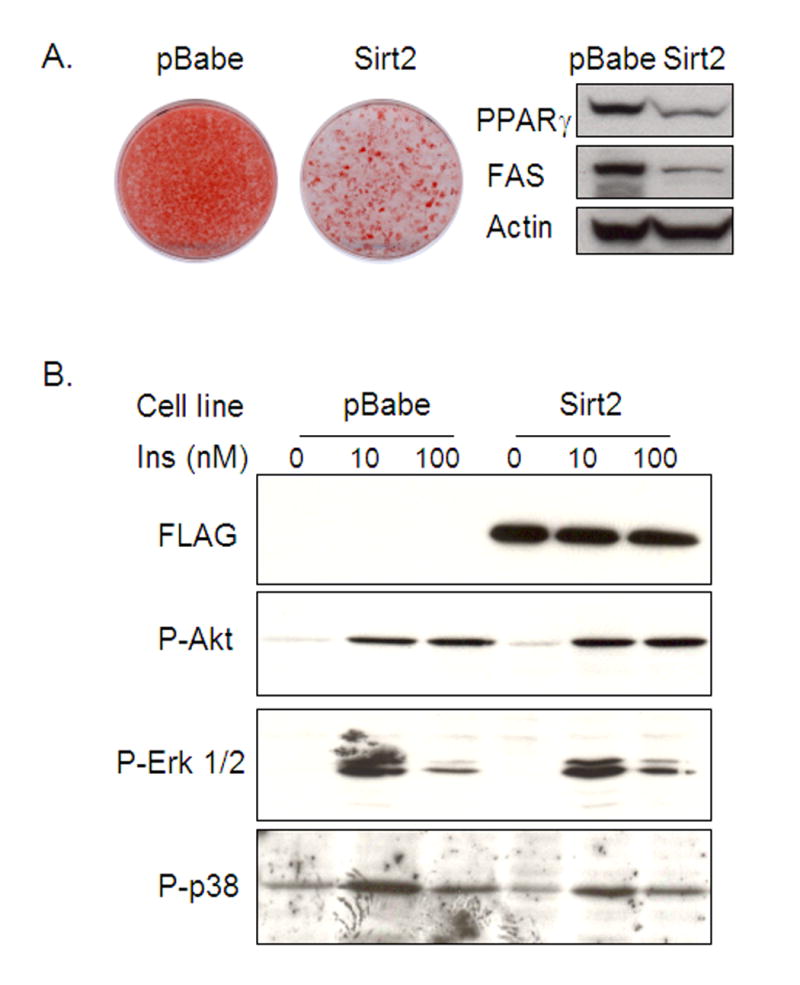
Sirt2 overexpression inhibits 3T3-L1 adipocyte differentiation without affecting insulin signaling in preadipocytes. (A). Following the differentiation protocol described in Material and Methods, Oil Red O staining of stably transfected 3T3-L1 cells with either control pBabe vector or Sirt2-FLAG-pBabe overexpression construct showed that exogenous Sirt2 overexpression inhibited adipocyte differentiation as compared with control cells. (B). Insulin signaling was assessed by western blotting of phospho-Akt, phospho-p38 and phospho-MAP kinase in confluent 3T3L1 preadipocytes. Stimulation was performed using 10 nM and 100 nM insulin for 5 min.
Sirt2 interacts with and deacetylates FoxO1 in 3T3-L1 preadipocytes
FoxO1, a known inhibitor of adipogenesis, has been previously shown to undergo regulated acetylation and deacetylation (Matsuzaki et al., 2005; Perrot and Rechler, 2005; Daitoku et al., 2004). Since there was no change in FoxO1 expression at the mRNA level, we explored whether FoxO1 protein expression or acetylation might be changed. Immunoprecipitation using anti-acetyl-lysine antibody followed by blotting with anti-FoxO1 antibody revealed that in control shGFP cells, most of the FoxO1 protein was in a deacetylated state, i.e. FoxO1 could not be detected in precipitated total acetylated protein. By contrast, in the Sirt2 knockdown cells, FoxO1 acetylation was markedly increased, and the anti-FoxO1 antibody easily detected the presence of FoxO1 protein in the precipitated lysate (Figure 4). This effect was specific because western blot analysis with anti-FoxO3a antibody did not detect any increased protein acetylation (data not shown). These effects on FoxO1 acetylation occurred with no change in the total level of FoxO1 protein in Sirt2 knockdown cells and no change in the level of Sirt1 protein(Figure 4), another member of the Sirt family which is able to deacetylate FoxOs.
Figure 4.
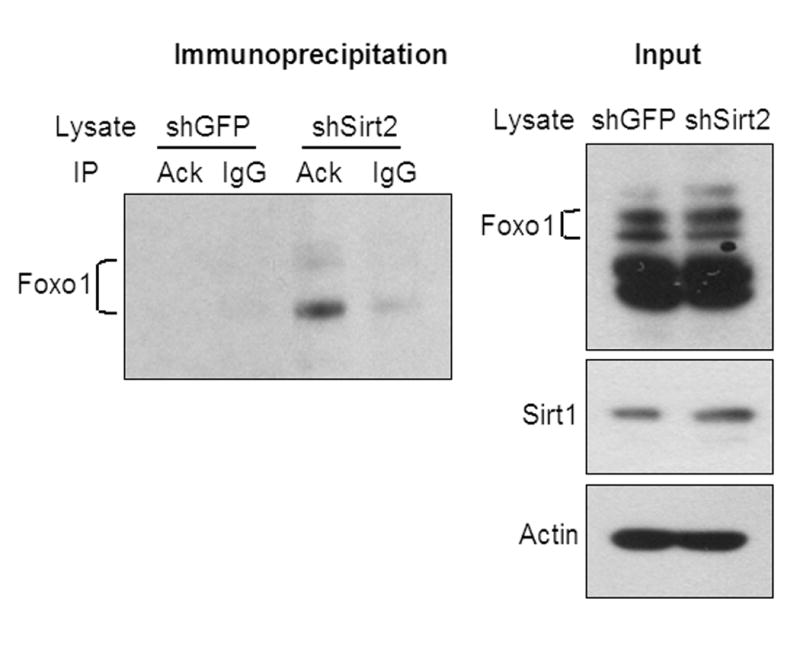
Sirt2 knockdown promotes FoxO1 acetylation. Non-denaturing total protein extracts from either shGFP or shSirt2 cells were immunoprecipitated with anti-acetylated-lysine antibody and precipitated lysates were blotted with anti-FoxO1 antibody. Total lysate input was detected by western blotting.
The increased acetylation on FoxO1 in Sirt2 knockdown cells indicates that FoxO1 can serve as a potential target for Sirt2 deacetylase activity. To investigate if Sirt2 interacts with FoxO1 directly, we performed immunoprecipitation of total cell lysates of cells overexpressing Sirt2-FLAG versus control cells infected with the empty pBabe retrovirus using a monoclonal anti-FLAG antibody conjugated to agarose. The immunoprecipitates were then immunoblotted with anti-FoxO1 antibody. In the cells expressing the Sirt2-FLAG construct, the anti-FLAG antibody co-precipitated significantly more FoxO1 protein than in control cells (Figure 5A) indicating that Sirt2 is present in a complex with FoxO1 protein. Western blot analysis of the same cell lysates with anti-FoxO1 antibody showed that this occurred with no difference in total FoxO1 protein content between Sirt2-FLAG and control cell lines (Figure 5A). The unchanged FoxO1 protein levels in preadipocytes of both Sirt2 knockdown and overexpressing cells is consistent with the realtime PCR data indicating that FoxO1 mRNA expression was not altered in Sirt2 knockdown cells during 3T3-L1 differentiation (Figure 2B). In lysates from cells overexpressing recombinant Sirt2-HA and FoxO1-FLAG, Sirt2-HA can co-immunoprecipitate FoxO1-FLAG in vitro (Figure 5B), confirming the interaction between Sirt2 and FoxO1 protein.
Figure 5.
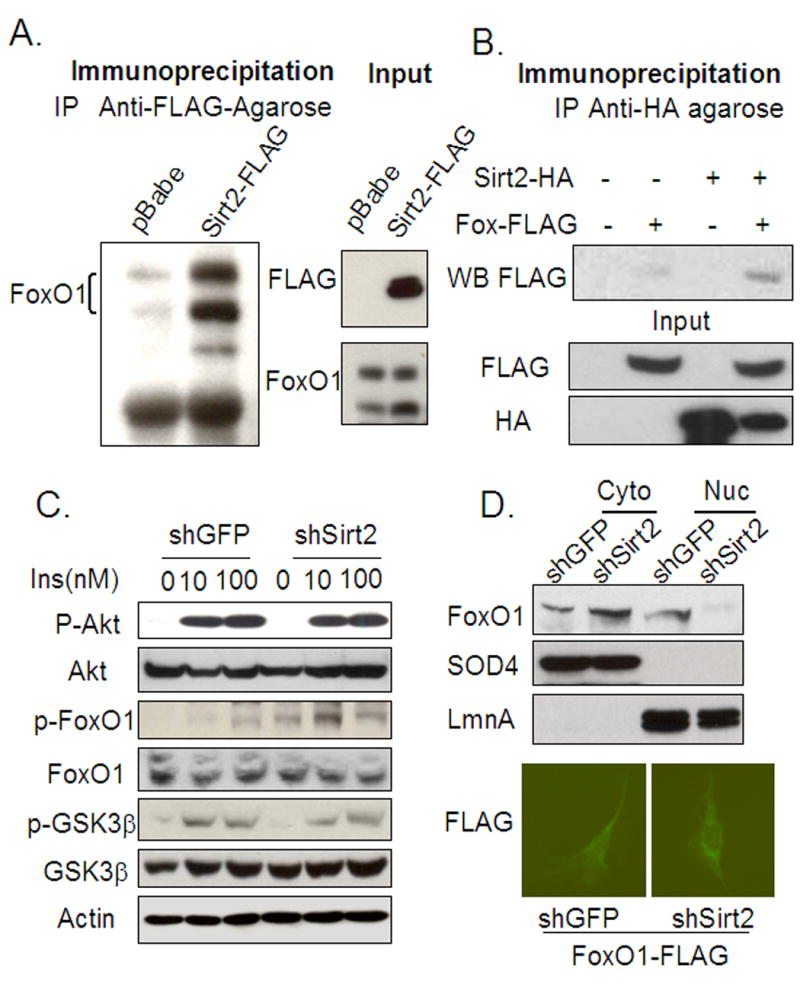
Sirt2 interacts with FoxO1 in vitro and Sirt2 knockdown promotes FoxO1 phosphorylation and cytosolic localization. (A) The non-denaturing lysates from either pBabe control or Sirt2-FLAG overexpression cell lines were immunoprecipitated with anti-FLAG-agarose. The precipitated lysates were blotted with anti-FoxO1 antibody. Markedly more FoxO1 protein was precipitated with anti-FLAG-agarose from Sirt2-FLAG overexpressing cells. (B) The non-denaturing lysates from HEK293 cells transiently transfected with Sirt2-HA and/or FoxO1-FLAG overexpressing constructs were subjected to immunoprecipitation with anti-HA agarose. Western blot of protein eluted from HA-Agarose shows that there is interaction between Sirt2 and FoxO1 in vitro. (C) The shGFP or shSirt2 cells were acutely (5 or 15 minutes) stimulated with different concentrations of insulin (10 nM and 100 nM) after serum deprivation. Insulin stimulated Akt and GSK3β phosphorylation (5 min stimulation) and FoxO1 phosphorylation (15 min stimulation) were assessed by western blotting. (D) Following a modified protocol for cytosolic and nuclear extract described previously (Emanuelli et al., 2000), lysates from both shGFP and shSirt2 cells were subjected to western blot analysis with anti-FoxO1 antibody. There was more FoxO1 protein translocated to the cytosol in shSirt2 3T3L1 cells. SOD4 and LaminA (LmnA) bands showed effective separation of nuclear and cytosolic proteins. Immunocytochemistry was done with cells carrying FoxO1-FLAG overexpressing construct with either stably transfected shGFP or shSirt2. Cells were fixed 48 hours after being plated in 10% FBS DMEM media. The anti-FLAG-FITC was used to detect subcellular localization of the recombinant FoxO1 in the cells.
Acetylation of FoxO1 regulates its phosphorylation and adipocyte differentiation in 3T3-L1 cells
To determine if the increased acetylation of FoxO1 could alter its ability to undergo phosphorylation, we treated serum-deprived shSirt2 and shGFP preadipocytes with insulin at different concentrations and immunoblotted cell extracts with an antibody that detects phosphorylation of FoxO1 Ser-253, the major site of FoxO1 phosphorylation by Akt/PKB (van der Heide et al., 2004). Consistent with the data above, insulin stimulated Akt/PKB phosphorylation to the same level in the Sirt2 knockdown and control cell lines. On the other hand, phosphorylation of FoxO1 on Ser-253 was increased two-fold in the Sirt2 knockdown cell line (Figure 5C). Since phosphorylation is known to affect nuclear translocation, nuclear and cytosolic extracts from shGFP and shSirt2 cells were prepared and subjected to immunoblot analysis with anti-FoxO1 antibody to. This revealed a 2- to 3-fold increase in the level of cytosolic FoxO1 protein in Sirt2 knockdown cells. Also, it was clear that the cytosolic FoxO1 band migrated in a slightly retarded position on the gel in the shSirt2 cells, consistent with increased FoxO1 phosphorylation, whereas the nuclear FoxO1 protein migrated at a lower position on the gels due to its unphosphorylated state. Furthermore, nuclear FoxO1 was decreased in amount (Figure 5D). Due to high background of FoxO1 antibody, we generated 3T3 L1 cell lines overexpressing FLAG tagged FoxO1, along with either shGFP or shSirt2 stable constructs, then used anti-FLAG- to detect the subcellular localization of recombinant FoxO1. FoxO1-FLAG was largely excluded from nucleus of cells overexpressing shSirt2, while cells overexpressing shGFP showed more diffusive pattern of FoxO1-FLAG localization (5D). Immunoblot of total protein lysates with anti-FoxO1 antibody revealed that no difference in total FoxO1 protein between the two lines (Figure 5C).
Analysis of FoxO1 phosphorylation using acetylation mutants
To further analyze the possible role of FoxO1 acetylation in regulation of FoxO1 phosphorylation, we used 3T3-L1 cell lines overexpressing either wild type FoxO1 or two FoxO1 mutants that mimic different acetylation states of the protein. In the KQ mutant, the three lysine residues surrounding Ser-253 known to be sites of acetylation, were replaced by glutamine residues. In the KR mutant, these lysines were replaced by arginine residues. All three overexpression constructs were generated with a N-terminal FLAG tags to allow quantitation of the protein. Immunoblotting of lysates from confluent cells overexpressing either FoxO1 wild type or the KQ and KR mutants with anti-FLAG monoclonal antibody revealed that all three proteins were equally overexpressed (Figure 6A). Quantitative PCR indicated a 5-fold increase in total FoxO1 mRNA in each line as compared to endogenous FoxO1 levels (Supplemental figure 2). Quantitative PCR using primers targeting the untranslated region of endogenous FoxO1 mRNA demonstrated that the endogenous FoxO1 expression level was not affected by expression of the exogenous protein (data not shown).
Figure 6.
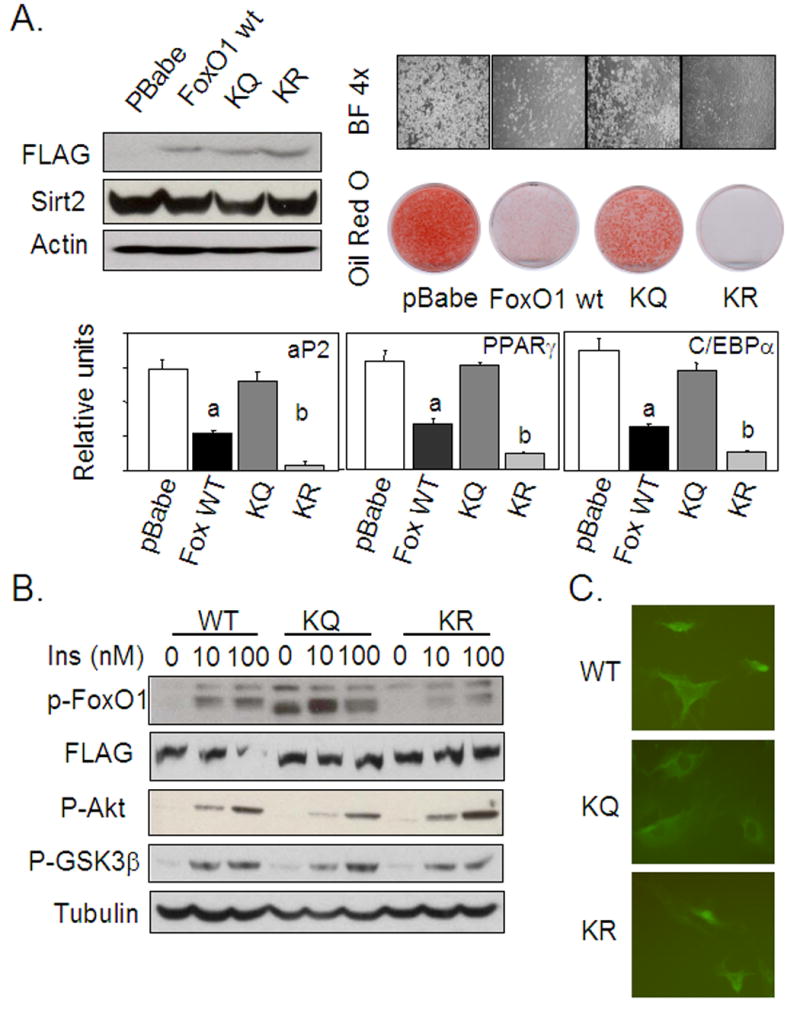
FoxO1 acetylation/deacetylation mimics regulate 3T3-L1 adipocyte differentiation and FoxO1 phosphorylation. (A) Different FoxO1 overexpression constructs were made with either wild type FoxO1 amino acid sequence or replacing all three lysine residues surrounding Ser-253 with Glutamine (KQ) or Arginine (KR). The FoxO1 WT, KQ, and KR overexpression constructs were all FLAG tagged. Overexpression was determined by western blotting using anti-FLAG antibody. Quantitative PCR with primers targeting the FoxO1 coding region showed that the level of overexpression of different constructs was similar and was about 5 times the level of endogenous FoxO1 observed in control cells (Supplemental figure2). Cell lines carrying various FoxO1 overexpression constructs or control cells were subjected to the differentiation protocol described in Material and Methods. Oil Red O staining of cells eight days after differentiation induction showed differences among different cell lines. Realtime PCR quantification of different adipocyte markers was consistent with the degree of adipocyte differentiation as accessed by bright light microscopic image and Oil Red O staining. Cells overexpressing wildtype FoxO1 had significantly decreased mRNA expression of different adipocyte differentiation markers comparing with control cells, as indicated with “a”; while cells expressing the KR mutant had significantly decreased mRNA expression compared with that of wild type FoxO1 overexpression, as indicated with “b”. (B) FoxO1 mutations mimicking different lysine acetylation states affect Ser-253 phosphorylation of FoxO1 and 3T3L1 adipocyte differentiation. After serum deprivation for 12 hours, 3T3L1 cells carrying wild type FoxO1, KQ, and KR mutant overexpression constructs were acutely stimulated with different concentrations (10 nM or 100 nM) of insulin for 10 minutes. Total cell extracts were subjected to western blot analysis to assess insulin stimulated phosphorylation status of FoxO1, Akt, and GSK3β. KQ mutant overexpression promoted both basal and insulin stimulated FoxO1 phosphorylation, whereas cells overexpressing the KR mutant had decreasedvFoxO1 phosphorylation in response to insulin, both as compared to cells overexpressing wildtype FoxO1. (C) Immunocytochemistry of different FoxO1 wildtype and mutants overexpressing cells was performed using anti-FLAG-FITC. Different subcellular localization patterns are observed for FoxO1 mutants.
The cell lines overexpressing wild type and mutant FoxO1 were subjected to the standard adipogenic differentiation protocol and stained with Oil Red O. Cells overexpressing wild type FoxO1 showed much less Oil Red O staining, consistent with a significantly decreased level of differentiation, than cells infected with the empty vector. This finding is consistent with known ability of FoxO1 to suppress adipogenesis. The cells overexpressing the KQ mutant of FoxO1, which mimics the acetylated state, exhibited enhanced differentiation compared with cells overexpressing wild type FoxO1. In contrast, cells whereas overexpressing the KR mutant, which mimics the deacetylated protein, showed decreased differentiation compared with cells overexpressing wild type FoxO1 (Figure 6A). These differences in lipid accumulation correlated well with expression of different adipocyte differentiation markers such as aP2, PPARγ, and C/EBPα by quantitative PCR (Figure 6A).
Assessment of FoxO1 Ser-253 phosphorylation after insulin stimulation in these cell lines revealed increased phosphorylation of the KQ mutant in the basal state, as well as a substantially higher level of phosphorylation following insulin stimulation when compared with cells overexpressing wild type protein. By contrast, cells expressing the KR mutant of FoxO1 showed decreased Ser-253 phosphorylation in the insulin-stimulated condition (Figure 6B). Thus, the FoxO1 acetylation mimic had increased Ser-253phosphorylation, whereas the deacetylated FoxO1 mimic had decreased Ser-253 phosphorylation. The FLAG western blot showed that the total recombinant FoxO1 protein expression is not altered under above conditions. These changes on FoxO1 phosphorylation occurred with no change in the level of phosphorylated/activated Akt and phosphorylated GSK3β (Figure 6B). The subcellular localization of FoxO1 mutants detected by immunocytochemistry using anti-FLAG-FITC was consistent with the observed differences in localization by sub-cellular fractionation and FoxO1 phosphorylation. Both FoxO1 wild type and KR overexpression had a diffused distribution within the cell, but KQ mutant had a nuclear exclusion pattern, where the KQ was more phosphorylated and localized in the cytoplasm (Figure 6B).
Discussion
The sirtuins represent a complex family of proteins that show homology to the yeast class III NAD-dependent protein/histone deacetylase Sir2. Sirt1, the best characterized member of the family, is localized mainly in the nucleus, and has been shown to interact with and regulate a number of transcription factors, corepressors, and coactivators thereby modulating downstream biological processes. Other Sirt proteins have different subcellular localizations, require different co-factors, and even have other enzymatic activities, suggesting that their biological functions are quite different (Liszt et al., 2005; North and Verdin, 2004). Sirt2, for example, is localized primarily in the cytoplasm and has deacetylase activity (Dryden et al., 2003; North et al., 2003), but its biological functions remain largely unknown.
In the present study, we find that Sirt2 mRNA is more abundant than other Sirts in both adipose tissue in vivo and preadipocytes in culture, with quantitative mRNA levels being four to seven times higher than those for Sirt1 or Sirt3. In addition, Sirt2 and Sirt1 expression is down regulated during adipocyte differentiation, whereas Sirt3 mRNA levels increases. By creating 3T3-L1 cell lines with stable overexpression and knockdown of Sirt2, we find that high levels of Sirt2 expression inhibit adipocyte differentiation, whereas reducing Sirt2 levels has the opposite effects. The promotion of adipocyte differentiation by Sirt2 is associated with increased expression of C/EBPα, PPARγ, Glut4, aP2, and FAS mRNAs, as well as increased expression of C/EBPβ, one of the earliest transcriptional changes in the normal program of adipocyte differentiation (Tang et al., 2004). Thus Sirt2 must act upstream of C/EBPβ at an even earlier event in induction of adipogenesis, and this appears to be at the level of FoxO1 acetylation/phosphorylation. Reducing the level of Sirt2 in the knockdown cells results in an increased level of FoxO1 acetylation, which in turn allows increased phosphorylation on Ser-253, excluding FoxO1 from the nucleus. This allows differentiation to progress, likely by reducing the ability of FoxO1 to interact with the PPARγ promoter and repress PPARγ transcription (Armoni et al., 2006). Although there is evidence that FoxO1 also acts during late stage differentiation, the effect of FoxO1 over-expression on differentiation appear to occur prior to the induction of early differentiation markers like C/EBPβ/σ, possibly at the level of clonal expansion. The effect of Sirt2 knockdown suggests that Sirt2 may act on FoxO1 during this clonal expansion stage.
In the process of adipocyte differentiation, insulin and/or IGF-1 act to stimulate FoxO1 phosphorylation on serine residues through activation of Akt. The serine phosphorylation of FoxO1 excludes it from the nucleus (Zhang et al., 2002), thus reducing its ability to repressing PPARγ transcription. Changing the level of Sirt2 alters the phosphorylation status of FoxO1, in this case not because of a change in insulin/IGF-1 action on Akt, but because phosphorylation of FoxO1 can also be regulated by acetylation/deacetylation of the lysine residues surrounding serine 253, the major site of regulatory phosphorylation (Zhang et al., 2002; Matsuzaki et al., 2005). While previous studies have suggested that CBP can act as a FoxO1 acetyl-transferase (Matsuzaki et al., 2005; Perrot and Rechler, 2005), it is not clear which enzyme deacetylates FoxO1. In the nucleus, Sirt1 has been shown to deacetylate FoxO1. This increases the level of FoxO1 localized in the nucleus, allowing it to be transcriptionally active (Frescas et al., 2005). In this study, we find that FoxO1 can also be a target of the cytoplasmic Sirt2 deacetylase, and that in this context Sirt2 plays a potentially important role in adipocyte differentiation.
There are several criteria by which this effect on differentiation appears to be the direct action of Sirt2 rather than an indirect effect of Sirt1. First, FoxO1 acetylation is increased by Sirt2 knockdown. This effect on FoxO1 acetylation is independent of changes in levels of Sirt1 or FoxO1 expression, supporting the notion that Sirt2 deacetylates FoxO1, rather than acting indirectly by decreasing Sirt1 expression level. Secondly, Sirt2 interacts with FoxO1 as shown by co-immunoprecipitation experiments. Third, in Sirt2 knockdown cells there is increased Ser-253 phosphorylation in response to insulin stimulation, and results in nuclear exclusion of FoxO1. This releases adipogenesis from FoxO1 inhibition. These data are consistent with the notion that acetylation of FoxO1 in the cytoplasm makes it more accessible to Akt phosphorylation, in turn promoting retention of FoxO1 in the cytosol, where it is transcriptionally inactive, and thus unable to repress expression of genes like PPARγ. In this way, increased acetylation reduces the inhibitory effect of FoxO1 on adipogenesis and promotes differentation.
This role of acetylation of FoxO1 in adipogenesis is further supported by our studies using FoxO1 mutants. There are three lysine residues surrounding the serine 253 in the wild type mouse FoxO1 protein. These three lysine residues can be acetylated by the protein acetyl-transferase CBP and deacetylated by Class III deacetylases, such as Sirt2. Recent studies have shown that acetylation/deacetylation of these lysines and serine phosphorylation can act in a synergistic manner (Matsuzaki et al., 2005). Thus when FoxO1 is acetylated by CBP, it is more accessible to phosphorylation, and this leads to its cytosolic translocation. In the FoxO1 KQ mutant, three lysine residues surrounding Ser-253 are replaced by glutamine. Previous studies on p53 have shown that substitution of glutamine for lysine serves to mimic a constitutive “acetylated” form of the p53 (Wang et al., 2003). On the other hand, replacing lysine with arginine, as in the FoxO1 KR mutant, serves to mimic the “deacetylated” form of protein (Feng et al., 2005; Marcotte et al., 2004). In agreement with Matsuzaki, et al (Matsuzaki et al., 2005), we find that these two FoxO1 mutants behave differently in terms of acetylation and Ser-253 phosphorylation in response to insulin stimulation when compared with wild type FoxO1. Thus, overexpression of the KR mutant, which is acetylation resistant, inhibits 3T3-L1 differentiation to an even greater extent than wild type FoxO1, whereas overexpression of the KQ mutant that mimics acetylated FoxO1 promotes differentiation. In each case, this correlates with the serine phosphorylation of the FoxO1 protein. Cells overexpressing WT FoxO1 exhibit an increased level of Ser-253 phosphorylation following insulin activation of Akt, while cells expressing the KQ mutant have higher levels of FoxO1 phosphorylation with or without any insulin stimulation. Cells overexpressing the KR mutant demonstrate the opposite with reduced FoxO1 phosphorylation following insulin stimulation. Since all these occur with the same level of Akt and GSK3β phosphorylation/activation, these findings indicate that it is an intrinsic property of FoxO1 and its apparent acetylation status that modulates FoxO1 phosphorylation and adipocyte differentiation. In addition, it is known that changes of FoxO1 lysine residue acetylation can affect its DNA binding activity. It is possible that the effects of FoxO1 mutants on adipocyte differentiation are mediated by similar changes. Combining these data with previous studies indicating that Sirt1 can also deacetylate FoxO1, it is possible that Sirt2 may target FoxO1 in the cytoplasm, while Sirt1 catalyzes FoxO1 deacetylation in the nucleus. It is also possible these two proteins recruit different co-factors and have different physiological or pathological regulation allowing them to carry out distinctive functions on the target. It is not known if there is any specificity of Sirt2 in deacetylation of specific lysine residues of the FoxO1 protein. It would also be of interest to compare the enzymatic activity of Sirt1 and Sirt2 on each potential acetylation site to further determine their relative roles on FoxO1.
While the current data support an important role of Sirt2 in adipocyte differentiation, how Sirt2 acts in on this process in normal physiology needs further study. Since many of the Class III HDACs of the sirtuin family require NAD as a cofactor, it is quite possible that in addition to the level of expression, one potential regulator of the activity of Sirt2 in normal cells is the level of NAD. This would allow Sirt2 to serve as a sensor of the cellular redox state and nutrient input with the ability to regulate gene expression and metabolism. It will be interesting to determine if Sirt2 can interact with FoxO1 in other tissues, such as liver, where FoxO1 has an important role in control of gluconeogenesis and its response to nutrient input and stress conditions.
Transcriptional activation and repression in eukaryotic cells has been shown to be involved closely with protein acetylation/deacetylation mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). The reversible acetyl-modification on lysine residues of transcription factors provides a mechanism by which modulating activities of either HATs or HDACs leads to changes in the expression of genes in metabolic pathways. This process could be further modulated by nutritional and redox state.
Sirt2, like Sirt1 is ubiquitously expressed, suggesting important functions in many types of cells. Undoubtedly, more Sirt2 targets await identification. It seems likely that Sirt2 will regulate other transcription factors besides FoxO1. By analogy with FoxO1, the acetylation/deacetylation of these factors may promote or antagonize their ability to undergo phosphorylation and regulate transcription. Drugs that activate or inhibit Sirt2 could regulate the acetylation state of FoxO1 and likely other transcription factors, thereby potentially regulating a wide variety of biological events. Sirt2 may serve as the key regulator for multiple targets, most of which are yet to be discovered.
Experimental Procedures
Cell culture and adipocyte differentiation
HEK293 cells and 3T3-L1 (American Type Culture Collection, ATCC, Manassas, VA) preadipocytes were cultured in high-glucose (400 mg/dl) Dulbecco’s modified Eagle medium (DMEM, Invitrogen) containing 10% fetal bovine serum (FBS) (Gemini Bioproducts). 3T3-L1 cells, including different stable transfected cell lines used for differentiation, were maintained in 10% FBS DMEM with high glucose. Differentiation was induced 2 days after the cells reached confluence (day 0) by adding an induction cocktail containing 100 nM insulin (Sigma), 1 μM dexamethasone (Dex) (Sigma-Aldrich), and 0.5 mM 1-methyl-3-isobutyl-xanthine (IBMX) (Sigma-Aldrich) to the medium containing 10% FBS. After 2 additional days (day 2), the medium was replaced by DMEM 10% FBS containing 100 nM insulin, and then media was changed every 2 days until the cells became mature adipocytes (day 10). All cells were maintained and differentiated at 37 °C in an environment with 5% CO2.
Plasmids and constructs
For overexpression, a Sirt2-FLAG and Sirt2-HA construct was prepared using Sirt2 cDNA derived from 3T3-L1 total cDNA produced by reverse transcription polymerase chain reaction, and inserted into pBabe-Bleo retroviral vector (Wei et al., 2003). Sirt2 shRNAs were designed using the Dhamarcon website. Oligos containing sense and antisense siRNA sequence with separating loop region were synthesized by IDT DNA Technology Inc. Oligo pairs were annealed in a buffer containing 100 mM Tris HCL (pH 7.5), 1 M NaCl, and 10 mM EDTA, and then inserted into HindIII-BglII sites of pSuper-Retro vector (McIntyre and Fanning, 2006; Taxman et al., 2006). Oligonucleotide sequences are shown in Supplemental Table 1.
The FLAG tagged wild type FoxO1, KQ (lysine residues converted to glutamine) and KR (lysine residues converted to arginine) mutants cDNA were gifts from Dr. Akiyoshi Fukamizu of University of Tsukuba, Japan. Constructs of FoxO1 wildtype and mutants for overexpression were subcloned into pBabe bleo retroviral vectors.
Immunoprecipitation and Western Blot Analysis
For immunoprecipitation experiments, cells were grown to confluence, non-denaturing cell lysates were prepared and immunoprecipitation was done as previously described (Entingh et al., 2003).
Western blot experiments were done after treatment and sample collection. Cell lysate was fractionated by SDS-10% polyacrylamide gel electrophoresis and transferred to PVDF membranes (Amersham). After blocking with recommended blocking reagents for 1 h at room temperature, the membranes were incubated overnight at 4°C with different antibodies. Antibodies used for western blot and IP are shown in Supplemental Table 2. The membranes were incubated with 1:2000-1:10000 secondary antibodies conjugated with HRP for 1 h at room temperature after washing for 10 minutes 3 times. Signals were detected by using the Amersham ECL chemiluminescence system and visualized by autoradiography.
Retroviral Infection and Transient Transfection
Retroviruses were produced as previously described (Entingh et al., 2003). Stable retroviral transduction of 3T3-L1 cells was achieved by infection for 12-16 hours. The cells were plated into 30 cm diameter Petri dishes and grown for 48-72 hours, after which selection with either Puromycin (2 μg/ml) or Zeocin (250 μg/ml) was initiated. Selection was stopped as soon as the non-infected control cell died off, and the media was replaced with normal growing media. The efficacy of overexpression was determined by western blot. The efficacy of knockdown at the protein level was assessed using both western blots.
The co-transfection for recombinant Sirt2-HA and FoxO1-FLAG was done in HEK293 cells using Lipofectamine2000 (Invitrogen).
Immunocytochemistry
After grown on coverslips for 48 hours in 10% FBS DMEM media, cells were fixed with 10% formalin, washed with PBS 3 times, then permeablized with 1% TritonX 100 and 1% BSA in PBS. After washing 3 times, fixed cells were blocked with 10% goat serum and 1% BSA for 1 hour, then incubated with FLAG- conjugate antibody in 1% BSA for 1-2 hours. Signal was detected using GFP fluorescent microscope..
Realtime Quantitative PCR
RNA samples were extracted using RNeasy kit (Qiagen). Each condition was performed in triplicate to allow for statistical analysis. The cDNA was synthesized using 1 μg total RNA using All Advantage RT-PCR kit. For quantification of relative expression levels of different Sirt mRNAs, 5 μl of cDNA was used for each reaction. To quantify the molar amount of RNA present in the samples, end product of realtime PCR for different Sirt genes were purified with PCR MiniElute kit (Qiagen), then quantified with NanoDrop 1000 and serially diluted 10-fold for each product, quantitative realtime PCR was performed using diluted PCR products with corresponding primers, Ct values of different dilutions were obtained, and linear regression graphs were created for each gene with absolute units derived from Ct values and corresponding molar amount based on PCR sizes. The corresponding target transcript molar amount used in Quantitative realtime PCR was accessed from the linear regression, then the molar amount of each gene per microgram total RNA was calculated based on total cDNA synthesis reaction volume and cDNA volume used for realtime PCR. For the differentiation time course experiments, realtime PCR was performed with 5 μl of cDNA using Sybrgreen master mix (Applied Biosystems) on ABI 7000 thermal cycler, and dCt values were collected by using either 18S ribosomal RNA or TATA-box binding protein (TBP) to normalize expression. The dCt values were calculated using absolute Ct values of the normalizer subtracted by Ct values of target genes. Final values were calculated using 2 exponential to the -dCt. Student t-test was performed between two different cell lines and significance was achieved when P<0.05.
Primers for realtime PCR using Sybrgreen are shown in Supplemental Table 3.
Supplementary Material
Acknowledgments
We thank Dr. Akiyoshi Fukamizu for providing cDNA constructs of different FoxO1 wild type and mutants, Dr. Brice Emanuelli and Dr. Jeremie Boucher for active discussion. We also thank Dr. Steven Russell for proofreading and comments about the manuscript.
The work was supported by research grant DK33201 to Dr. C. Ronald Kahn.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armoni M, Harel C, Karni S, Chen H, Bar-Yoseph F, Ver MR, Quon MJ, Karnieli E. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. 2006;281:19881–91. doi: 10.1074/jbc.M600320200. [DOI] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–9. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–7. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–85. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli B, Peraldi P, Filloux C, Sawka-Verhelle D, Hilton D, Van Obberghen E. SOCS-3 is an insulin-induced negative regulator of insulin signaling. J Biol Chem. 2000;275:15985–91. doi: 10.1074/jbc.275.21.15985. [DOI] [PubMed] [Google Scholar]
- Entingh AJ, Taniguchi CM, Kahn CR. Bi-directional regulation of brown fat adipogenesis by the insulin receptor. J Biol Chem. 2003;278:33377–83. doi: 10.1074/jbc.M303056200. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–95. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–80. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–95. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N. FOXO transcription factors in cell-cycle regulation and the response to oxidative stress. Antioxid Redox Signal. 2005;7:752–60. doi: 10.1089/ars.2005.7.752. [DOI] [PubMed] [Google Scholar]
- Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–4. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun. 1999;266:677–83. doi: 10.1006/bbrc.1999.1885. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Marcotte PA, Richardson PL, Guo J, Barrett LW, Xu N, Gunasekera A, Glaser KB. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal Biochem. 2004;332:90–9. doi: 10.1016/j.ab.2004.05.039. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–83. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre GJ, Fanning GC. Design and cloning strategies for constructing shRNA expression vectors. BMC Biotechnol. 2006;6:1. doi: 10.1186/1472-6750-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–29. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol Endocrinol. 2005;19:2283–98. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys. 2000 Spring;32:187–204. doi: 10.1385/cbb:32:1-3:187. [DOI] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–70. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Zhang JW, Daniel Lane M. Sequential gene promoter interactions by C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem Biophys Res Commun. 2004;318:213–8. doi: 10.1016/j.bbrc.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–45. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Taxman DJ, Livingstone LR, Zhang J, Conti BJ, Iocca HA, Williams KL, Lich JD, Ting JP, Reed W. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide LP, Smidt MP. Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem Sci. 2005;30:81–6. doi: 10.1016/j.tibs.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang YH, Tsay YG, Tan BC, Lo WY, Lee SC. Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem. 2003;278:25568–76. doi: 10.1074/jbc.M212574200. [DOI] [PubMed] [Google Scholar]
- Wei W, Jobling WA, Chen W, Hahn WC, Sedivy JM. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol Cell Biol. 2003;23:2859–70. doi: 10.1128/MCB.23.8.2859-2870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gan L, Pan H, Guo S, He X, Olson ST, Mesecar A, Adam S, Unterman TG. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277:45276–84. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


