Summary
Groucho-related (Gro/TLE/Grg) corepressors meditate embryonic segmentation, dorsal-ventral patterning, neurogenesis, and Notch and Wnt signaling. Although Gro/TLE/Grgs disrupt activator complexes and recruit histone deacetylases (HDAC), activator complexes can be disrupted in various ways, HDAC recruitment does not account for full corepressor activity, and a direct role for Gro/TLE/Grg binding and altering chromatin structure has not been explored. Using diverse chromatin substrates in vitro, we show that Grg3 creates higher order, condensed complexes of polynucleosome arrays. Surprisingly, such complexes are in an open, exposed configuration. We find that chromatin binding enables Grg3 recruitment by a transcription factor and the creation of a closed, poorly accessible domain spanning 3−4 nucleosomes. Targeted recruitment of Grg3 blankets a similar sized region in vivo, impairing activator recruitment and repressing transcription. These activities of a Groucho protein represent a newly discovered mechanism which differs from that of other classes of corepressors.
Keywords: Grg, groucho, FOXA, HES, nucleosome, chromatin, repression
Introduction
Transcriptional corepressors can be divided into classes by their mechanism of gene silencing. Upon recruitment of the Sir corepressors to a genomic target site, Sirs spread long distances on chromatin to stably repress gene activity (Pirrotta and Gross, 2005). Lengths of Sir-bound chromatin assume distinctive chromatin structures (Weiss and Simpson, 1998) that are resistant to nucleases (Loo and Rine, 1994), but not to access by activators, TBP, and Pol II (Sekinger and Gross, 2001). By contrast, the Polycomb class of corepressors do not spread from recruitment sites (Ringrose et al., 2004), but rather link distal sites together by apparent looping (Lavigne et al., 2004), whereupon they antagonize local chromatin remodeling (Francis et al., 2001) without generating nuclease-resistant structures (Dellino et al., 2004). In yeast, the Ssn6-Tup1 class can interfere with TBP binding and interactions between locally bound activators and the basal transcriptional machinery, but the mechanism of interference is unknown (Kuras and Struhl, 1999; Smith and Johnson, 2000). The Tup1 subunit contains two histone-binding regions (Edmondson et al., 1996) and Ssn6-Tup1 serves as a platform for recruiting histone deacetylases (HDACs) to chromatin (Watson et al., 2000). Despite that Ssn6-Tup1 globally regulates yeast genes involved in cell type control (Keleher et al., 1992), the intrinsic ability of the purified corepressor to bind and modulate chromatin has been unexplored.
Grg, TLE, and its founding family member Groucho (Gro), are metazoan transcriptional corepressors that are structurally and functionally related, and similar to yeast Tup1. Gro/TLE/Grg proteins mediate embryonic segmentation, dorsal-ventral patterning, neurogenesis, and Notch and Wnt signaling (Chen and Courey, 2000). In mammals, the Gro/TLE/Grg family consists of four unlinked genes, termed TLE1−4 in humans and Grg1−4 in mice. Gro/TLE/Grg proteins contain an amino-terminal tetramerization domain, a central regulatory domain with a nuclear localization signal, and a large carboxy-terminal domain (C-term) containing WD40 repeats that is highly conserved with Tup1 (Jennings et al., 2006).
Gro/TLE/Grg itself does not have a DNA binding domain; like Tup1, Gro/TLE/Grg is recruited to specific DNA sites by its binding partners. For example, recruitment of Gro/TLE/Grg by Hes1 represses neurogenic genes (Paroush et al., 1994), recruitment by TCF represses genes that are derepressed by Wnt signaling (Cavallo et al., 1998), and recruitment by PRDI-BF1/Blimp-1 represses the β-interferon promoter (Ren et al., 1999). Gro/TLE/Grg binding partners now include FoxD3 (Yaklichkin et al., 2007) and the endodermal transcription factor FoxA (Wang et al., 2000).
Like Tup1, Gro/TLE/Grg proteins can repress transcription by inhibiting the basal transcriptional machinery (Yu et al., 2001) and recruiting HDACs (Chen et al., 1999). However, HDAC activity only contributes partially to corepressor function in chromatin (Chen et al., 1999). Since Gro/TLE/Grg proteins interact with the tails of core histones H3 and H4 (Palaparti et al., 1997; Flores-Saaib and Courey, 2000), we speculated that they might have an intrinsic ability to bind, modulate, and protect chromatin to facilitate transcriptional repression.
To investigate this issue, we assessed the ability of a purified Gro/TLE/Grg protein to bind and modify the accessibility of various chromatin substrates. We further assessed the impact of specific DNA site recruitment of a Gro/TLE/Grg protein by FoxA1 and Hes1. We found that Grg3 stably binds purified nucleosome arrays and undergoes an apparent conformational change in the process. Unexpectedly, such chromatin binding enables target site recruitment by the transcription factors; recruitment does not occur on free DNA. The combination of chromatin and transcription factor binding, but not chromatin binding by Grg3 alone, enables the formation of a local domain of nucleosomes that is resistant to various nuclease probes; a similar local domain is blanketed by Grg3 recruitment in vivo, impairing activator access and causing transcriptional repression. The results reveal an intrinsic chromatin structure modifying function for the Groucho class of corepressor proteins.
Results
Grg3 interacts with chromatin at several levels
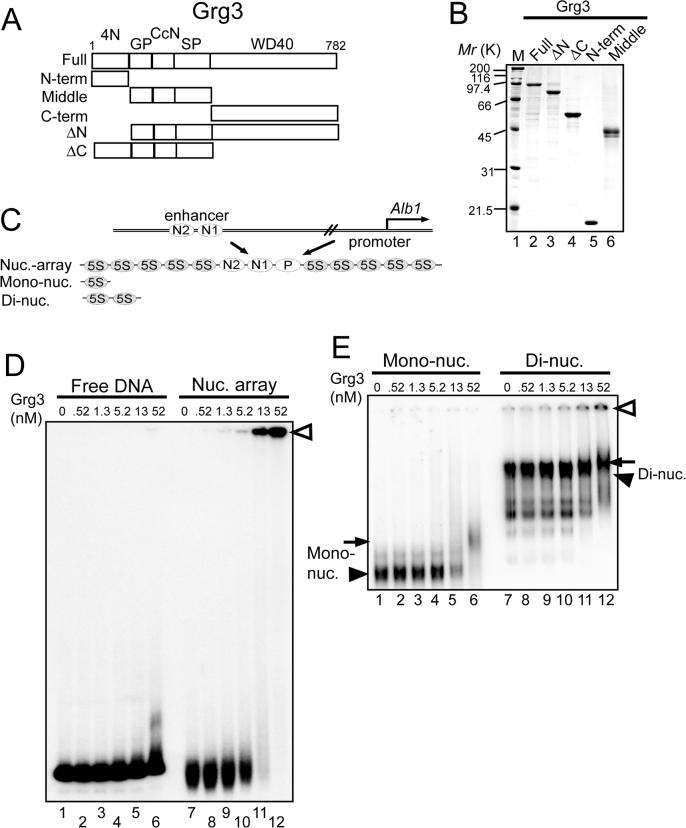
To investigate the ability of Gro/TLE/Grg proteins to interact with chromatin and modulate structure, we generated recombinant, purified Grg3 protein (Fig. 1A; 1B, lane 2) and assessed its binding to nucleosome arrays. The arrays were generated from end-labeled DNA templates and recombinant core histones; they contained 13 regularly spaced nucleosomes with a segment of the alb1 enhancer in the central “N1” nucleosome, which binds FoxA1 (Cirillo et al., 2002) (Fig. 1C). In electromobility shift assays (EMSA), Grg3 did not bind efficiently to the free DNA used to make the arrays, requiring 52 nM to begin to exhibit binding to 1 nM DNA (Fig. 1D, lanes 1−6). However, binding to 1 nM nucleosome arrays was detected at 5.2 nM Grg3 and was complete (loss of free array band) by 52 nM (Fig. 1D, lanes 10−12). FPLC gel filtration analysis showed that our recombinant Grg3 was predominantly a tetramer, whereas an N-terminally deleted Grg3 (Fig. 1B), lacking the tetramerization domain (Chen et al., 1998), was a monomer (Suppl. Fig. 1A-D). Thus, we observed quantitative array binding at ∼3 wild type Grg3 tetramers per array. Grg3 did not bind to arrays made with tailless histones H3 and H4, whereas binding by FoxA1 to the tailless arrays, as a control, was normal (Supp. Fig. 1E-H). Interestingly, the interaction between Grg3 and arrays resulted in higher order complexes that were retained in the wells (Fig. 1D, arrowhead). The data show that Grg3 alone is sufficient to preferentially bind arrays over free DNA, in a histone-tail dependent manner, and create higher order complexes.
Fig. 1. Grg3 binding to nucleosomal substrates.
(A) Deletion mutant series of Grg3. 4N, tetramerization domain; GP, glycine/proline rich; CcN, phosphorylation sites, nuclear localization; SP, serine/proline rich; WD40, repeat domain. (B) SDS-PAGE of purified Grg3 proteins. (C) alb1 gene and nucleosome templates (McPherson et al., 1993; Shim et al., 1998; Cirillo and Zaret, 1999). 5S: 5S rDNA. (D) EMSA with end-labeled alb1 enhancer-containing nucleosome arrays or free DNA; unbound DNA or nucleosome arrays at bottom of agarose gel. Complexes of Grg3 and arrays were retained in wells; open arrowheads. (E) EMSA with 5S rDNA mono- and dinucleosomes; polyacrylamide gel. Unbound probes; black arrowheads. Grg3 complexes with nucleosomal DNA, black arrows. The Grg3/dinucleosome complex was also detected at the wells; open arrowhead.
To test whether Grg3 interacts with mononucleosomal subunits, we performed EMSA with reconstituted mononucleosomes at 13 nM. Grg3 at 52 nM (13 nM tetramers) quantitatively bound the mononucleosomes (Fig. 1E, lanes 1−6). This was a markedly higher affinity than Grg3 binding to the free DNA for the arrays (compare Fig. 1D and 1E, lanes 6) or free DNA used for the mononucleosomes (data not shown), revealing an intrinsic affinity for the nucleosome core particle. However, Grg3 did not cause retention of mononucleosomes in the wells.
We speculated that retention in the wells was due to a structure uniquely assumed by a run of nucleosomes. Such a determinant could be assumed at the level of a long polynucleosome array, e.g., a solenoid, or merely at the level of a dinucleosome. We therefore performed EMSA with dinucleosomes at 6.5 nM. As with mononucleosomes, binding saturation began at 52 nM Grg3, but now was accompanied by the retention of the Grg3-dinucleosome complexes in the well (Fig 1E, lanes 7−12). We conclude that Grg3 has an intrinsic affinity to chromatin over free DNA that is mediated by histone tail binding and can result in a higher-order complex that depends upon at least a dinucleosomal subunit.
Grg3 condenses nucleosome arrays without preventing protein accessibility
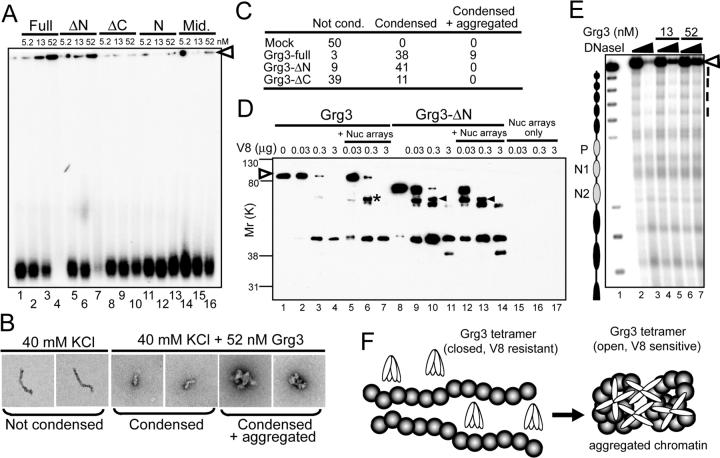
We next tested Grg3 domains required for chromatin binding. Grg3-full showed the strongest affinity to nucleosome arrays (Fig. 2A, lanes 1−4) and deleting the N-term tetramerization domain (Fig. 1A, B) reduced binding slightly (Fig. 2A, lanes 5−7). By contrast, deleting the WD40 domain (ΔC) greatly reduced binding, and isolated fragments of the N-term and middle regions did not bind at all (Fig. 2A, lanes 8−16). The isolated WD40 domain could not be produced. In summary, the WD40 domain most strongly contributed to chromatin binding.
Fig. 2. Condensation and aggregation of nucleosome arrays by Grg3.
(A) EMSA of designated Grg3 variant proteins and arrays. Arrowhead, complexes retained in the wells. (B) Electron microscopic analysis. Complexes were classified into categories indicated in (C). 50 arrays were analyzed for each Grg3 protein. (D) Conformation change of Grg3 upon interaction with arrays. Full- and ΔN-Grg3 proteins were digested with V8 protease and the Grg3 products were detected by Western blotting. Association of Grg3 with arrays induced V8 cleavage (asterisk), which is also detected with ΔN-Grg3 regardless of array addition (arrowhead). Open arrowhead, undigested Grg3. (E) DNaseI digestion of Grg3 binding to the arrays. Positions of nucleosomes on reconsitituted arrays are indicated to left of the gel. N1, N2, and P correspond to the alb1 enhancer N1, N2 sequences, and minimal promoter, respectively. Digestion was with 35 or 70 ng DNaseI. (F) Upon binding to chromatin Grg3 changes its conformation, and now both condenses chromatin and forms aggregates.
To evaluate the structure of Grg3-bound arrays, we used electron microscopy. Interestingly, Grg3 dramatically condensed the length of the arrays and often generated aggregates containing multiple arrays (Fig. 2B, C). Grg3-ΔN also induced array condensation; however, it could not generate aggregates (Fig. 2C). These data suggest that tetramerization of Grg3 is important for nucleosome array aggregation. Since both Grg3-full and -ΔN caused the arrays to be condensed and retained in the well, in EMSA, but only Grg3-full caused aggregation, well retention in EMSA correlates with array condensation and not aggregation. The WD40-deleted Grg3 was unable to elicit array aggregation and weakly condensed the arrays (Fig. 2C), consistent with its weak affinity to the arrays in EMSA (Fig. 2A, lane 10).
To analyze the mechanism of Grg3-mediated array condensation, we investigated the accessibility of Grg3 to protease digestion in the presence and absence of nucleosome arrays. We reproducibly observed a V8-sensitive domain in full-length Grg3 only in the presence of nucleosome arrays (Fig. 2D, lanes 1−7, asterisk). V8-sensitivity was not induced by free DNA (data not shown). Based on the size of the V8-generated product, the cleavage appears to be in the middle, regulatory domain of Grg3. Interestingly, a band at apparently the same gel position, along with a lower band, was observed with Grg3-ΔN regardless of nucleosome array interaction (Fig. 2D, lanes 8−14). Thus, binding to chromatin helps expose a V8 cleavage site in wild type Grg3 that is already exposed in the tetramerization-deficient mutant, in the absence of chromatin. These data suggest that Grg3 undergoes a conformational change at the level of subunit interactions upon binding chromatin (Fig. 2F and Discussion).
Given that Grg3 causes nucleosome arrays to both condense and aggregate, we anticipated that Grg3 would markedly affect nucleosome accessibility. Contrary to these expectations, in repeated assays Grg3 only modestly inhibited DNase I cleavage, as indicated by a small increase in the undigested fraction of the arrays and an increase in less-digested products (Fig. 2E, lanes 4−7, dotted line at side of gel; Fig. 4B). Thus, although Grg alone can strongly condense nucleosome arrays, it does not markedly exclude access by other proteins.
Fig. 4. Protection of 3−4 local nucleosomes by FoxA1 recruitment of Grg3.
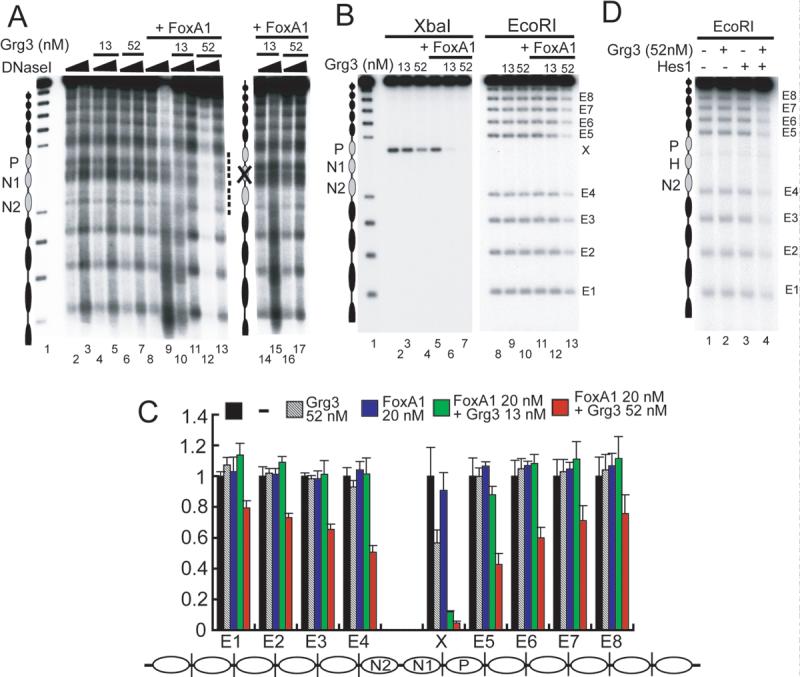
(A) DNaseI digestion (35 or 70 ng) of Grg3 and FoxA1 binding to arrays. Lanes 14−17, FoxA1 binding site mutant arrays. (B) Accessibility of restriction enzymes to arrays with or without Grg3 and FoxA1. Arrays were digested with either XbaI or EcoRI with or without Grg3 and 20 nM FoxA1. Each band (E1−8, X) labeled on right is shown in (C). (C) Quantification of the results from 3 experiments as in (B). Results of arrays alone were normalized to a value of 1 (black bars), and relative cleavage in the presence of proteins, at each site, is shown (colored bars). (D) EcoRI digestion of Grg3 and Hes1 binding to the Hes1-arrays. Arrays were digested with EcoRI with 52 nM of Grg3 (lanes 2, 4) and 40 nM of Hes1 (lanes 3, 4). The position of the nucleosome with Hes1 binding sites is denoted by 'H'.
The N-terminal region of Grg3 interacts with the C-terminal region of FoxA1
The inability of Grg3 alone to dramatically alter chromatin access led us to investigate transcription factors that could recruit Grg3. We previously discovered that FoxA1 protein could bind its target sites on condensed arrays in vitro (Cirillo et al., 2002) as well as on nucleosomes in vivo (Chaya et al., 2001). This is consistent with the apparent pioneer transcription factor activity of FoxA proteins in activating genes early in development (Bossard and Zaret, 2000; Lee et al., 2005). However, cofactors that work with FoxA are not well established. We therefore focused on a reported interaction between FoxA2 and a human homolog of Grg (Wang et al., 2000).
We covalently linked purified, full-length FoxA1 and FoxA2 to Sepharose beads and assessed the proteins' abilities to bind in vitro-translated (IVT) Grg3 and fragments thereof (Supp. Fig. 2A-C). Both Fox A1 and FoxA2 bound to full-length Grg3, but not to the isolated middle domain of Grg3 and only weakly to the C-terminal domain of Grg3 (Supp. Fig. 2C, lanes 5 and 7), though the latter was the previously reported FoxA2 interaction domain (Wang et al., 2000). Both FoxAs bound to Grg3-ΔC, but neither bound to Grg3-ΔN. Interestingly, both FoxAs bound most strongly to the isolated N-term region of Grg3. Because such binding was stronger than to Grg3 and Grg3-ΔC, the middle region of Grg3 appears to inhibit interactions between the N-term domain and FoxA.
We also found that FoxA1-full and -ΔN, but not -ΔC, efficiently bound recombinant, purified Grg3 (Supp. Fig. 2D, lanes 4−11). This result confirmed that the C-term region of FoxA proteins interacts with Grg proteins (Wang et al., 2000). We also tested full length Hes1, a prototype Gro/TLE/Grg recruiting factor, and a WRPW Hes1 deletion (Fisher et al., 1996), as positive and negative controls for stable Grg3 binding, respectively (Supp. Fig. 2A, B). As expected, Hes1-full efficiently bound to Grg3, whereas ΔWRPW failed to bind (Supp. Fig. 2D, lane 12−15).
FoxA1 and Hes1 recruit Grg3 to chromatin, but not to free DNA sites
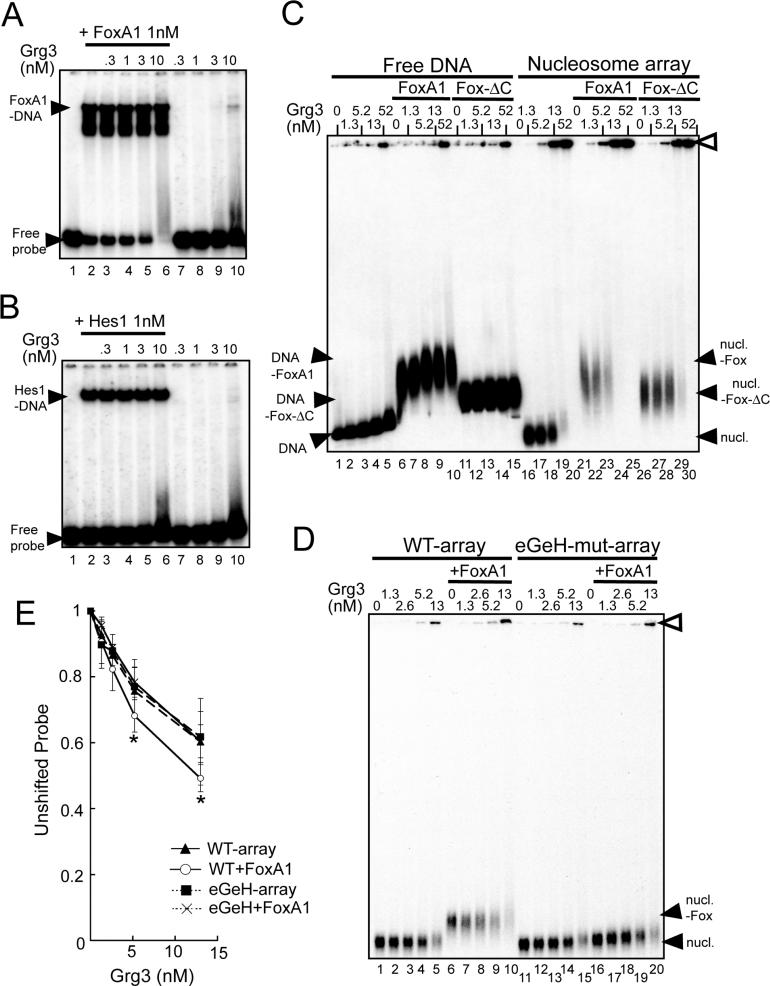
Since FoxA1 is able to bind its target site in compacted nucleosome arrays and create a locally open structure (Cirillo et al., 2002), we anticipated that it would be capable of recruiting Grg3 to chromatin in vitro. Similarly, we assumed that since Grg3 binds the C-term domain of FoxA1, outside the DBD, FoxA1 could recruit Grg3 to free DNA. Contrary to the latter expectations, in an EMSA we were unable to observe a Grg3-FoxA complex on a high-affinity free DNA binding site for FoxA (Fig. 3A; note absence of 'supershifted' complexes). The inability to observe ternary complexes was also seen with the 2.5 kb free DNA used for nucleosome arrays (Fig. 3C, lanes 7−10), even though FoxA1 at 5.2 nM quantitatively shifted the arrays (Fig. 3C, lane 6). We conclude that FoxA1 is unable to recruit Grg3 to a free DNA site. Similarly, we found that Hes1 is unable to recruit Grg3 to a high affinity Hes1 target on free DNA (Fig. 3B).
Fig. 3. FoxA1 recruitment of Grg3 to arrays, but not free DNA.
(A), (B) Absence of FoxA1-Grg3 and Hes1-Grg3 complexes on free DNA. End-labeled probes containing a FoxA1 (Liu et al., 1991) or Hes1 binding site (Lee et al., 2001) were incubated with FoxA1 or Hes1 (lane 2), Grg3 (lanes 7−10), or both Grg3 and FoxA1 or Hes1 (lanes 3−6). Note the absence of supershifting of FoxA1-or Hes1-DNA complex by Grg3. (C, D) FoxA1 enhances Grg3-array interactions, but not Grg3-DNA interactions (see text). The positions of unshifted DNA, arrays, and complexes are noted by black arrowheads. Complexes with Grg3 were primarily at wells (open arrowhead). (E) Quantitation of PhosphorImager data as in (D). n=4, *P < 0.05 for FoxA1 and Grg3 with wt arrays vs with eGeH site mutant arrays.
By contrast, FoxA1 appeared to enhance Grg3 binding to arrays (Fig. 3C, compare lanes 18, 19 with 23, 24). FoxA1 lacking its Grg3 interacting domain shifted the arrays, but did not enhance Grg3 binding (Fig. 3C, lanes 26−30 compared to 21−25). To investigate this further, we assessed a narrower range of Grg3 and compared the results to arrays containing mutations of FoxA binding sites (Fig. 3D, lanes 11−20, Supp. Fig. 1H). As quantified in Fig. 3E (open circles; p<0.05), we could detect FoxA1 significantly enhancing Grg3 binding to the wild type arrays, but not to the mutant arrays, despite that FoxA solely binds the middle nucleosome and Grg3 can independently bind any of the other 12 nucleosomes in the arrays (Fig. 1C, E). In summary, FoxA recruits Grg3 to chromatinized DNA, but not free DNA.
FoxA1 and Hes1 recruitment of Grg3 mediates the formation of locally closed, resistant chromatin
We next used nuclease probes to assess the consequences of enhanced Grg recruitment to nucleosome arrays. As before, Grg3 alone did not induce a marked change in the DNaseI digestion pattern, and FoxA1 binding alone somewhat enhanced nuclease sensitivity (Fig. 4A, lanes 1−9). However, adding both proteins induced protection against DNaseI digestion centering on the N1 nucleosome, which contains two FoxA binding sites (Fig. 4A, lanes 10, 11; compare with lanes 4, 5). At higher Grg3 concentrations, DNase protection extended to neighboring nucleosomes (Fig. 4A, lanes 12, 13). Protection was due to recruitment via the FoxA sites, because array templates with point mutations of the FoxA sites failed to exhibit the effect (Fig. 4A, lanes 14 through 17). Furthermore, Grg3-ΔN, which is impaired in FoxA1 interactions, also failed to induce protection on the wild type arrays in the presence of FoxA1 (data not shown). These results show that Grg3 is recruited to the central N1 nucleosome first by binding FoxA1, protecting that nucleosome, and then extending a resistant chromatin structure to a few neighboring nucleosomes. Transcription factor-recruited Grg3 is thus far more effective in creating a locally resistant chromatin structure than Grg3 alone.
When digesting the arrays with the restriction enzyme XbaI, whose unique target site is located in linker DNA near the N1 nucleosome, Grg3 alone inhibited XbaI digestion by about 50% and FoxA1 had little effect (Fig. 4B, lanes 3−5). However, FoxA1 and Grg3 together completely protected the XbaI site (Fig. 4B, lanes 6, 7; quantitated in 4C), despite that XbaI cleavage of the same free DNA is not inhibited by the proteins (Supp. Fig. 3). More strikingly, Grg3 alone had virtually no effect on EcoRI digestion of the linker DNA between the 5S rDNA repeats of the array, whereas FoxA1 and 52 nM Grg3 elicited strong protection at the Eco site nearest to the FoxA1 binding sites (Fig. 4B, lane 13, band E5; 4C). Both proteins together elicited protection of EcoRI sites extending 1−2 nucleosomes beyond N1 (Fig. 4B, C). Because 13 nM Grg3 with FoxA1 protected only the XbaI site, Grg3 recruited there appeared to nucleate further Grg3 recruitment, at 52 nM, and local extension of a nuclease-inhibitory structure. We conclude from the above studies with different nuclease probes that FoxA1 recruitment of Grg3 generates a nuclease-inhibitory structure over about 3−4 local nucleosomes.
To assess the generality of these findings, we investigated whether Hes1 can recruit Grg3 and induce the creation of a locally resistant structure. We constructed a nucleosome array which has 53 bp spanning the FoxA1 sites substituted for 53 bp from the Hes1 promoter, which contains two Hes1 binding sites (Takebayashi et al., 1994) (Supp. Fig. 3A). Both the Hes1-full and ΔWRPW bound specifically to the Hes1-array and not to the original albumin-array, whereas FoxA1 bound much stronger to the albumin-array (Supp. Fig. 4B). Notably, arrays incubated with Hes1 and Grg3 exhibited protection from EcoRI digestion at the site nearest to the Hes1 binding sites, extending to several nucleosomes (Fig. 4D). We conclude that creation of a locally compact, resistant chromatin structure could be a general property of Grg3 recruitment by a transcription factor. Recruitment could increase the local concentration of Grg3, thereby enhancing the modest effect that Grg3 alone has in chromatin compaction.
Grg3 represses FoxA1 target genes
Next, we investigated the biological consequences of Grg3 recruitment by FoxA1. An anti-pan-Grg antibody revealed that Grgs were expressed in 293T, CHO, and COS7 cell lines, but interestingly, not in any of liver cell lines, including Huh7, HepG2, BW1J, and H2.35 (Fig. 5A). Therefore, we used the liver lines to determine whether exogenous Grg3 affects the expression of endogenous FoxA target genes. We first used the temperature-sensitive H2.35 mouse line, which is maintained in an undifferentiated state at the permissive temperature (33° C) and expresses various liver genes at the restrictive temperature (37° or 39° C) (Zaret et al., 1988). We infected the cells with retroviruses expressing either full-length Grg3, Grg3-ΔN, or a GFP control protein (Fig. 5B) and evaluated the changes in expression of the well-established FoxA target genes alb1, afp, and ttr (Costa et al., 1989; Liu et al., 1991; Millonig et al., 1995) by qRT-PCR (Fig. 5C). The temperature shift induced these genes ∼60, ∼10, and ∼15 fold, respectively, in uninfected cells (Fig. 5C, points 1, 5; 9, 13; and 17, 21). Notably, transduction of full-length Grg3 impaired the induction about 5-fold for alb1 and 2−3 fold for afp and ttr (Fig. 5C, points 7, 15, 23). GFP or the Grg3-ΔN, lacking the tetramerization and FoxA interaction domain, had no effect (Fig. 5C, points 6, 8, 14, 16, 22, 24). We speculate that the absence of endogenous Grg expression in the liver cell lines is explained by the inhibitory effect of Grg on hepatocyte differentiation.
Fig. 5. Grg3 inhibits FoxA target gene expression in mouse and human cells.
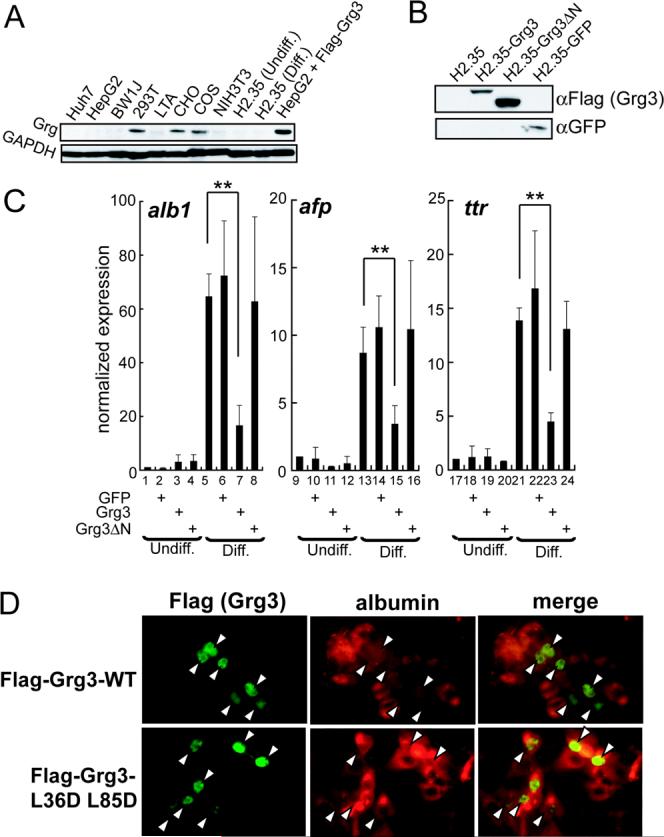
(A) Grg protein expression in cell lines was analyzed by Western blotting with the indicated antibodies. (B) Western blotting of whole cell extracts of H2.35, infected with Flag-Grg3-full, Flag-Grg3-ΔN, and GFP retroviruses as shown. (C) Effect of full- and ΔN-Grg3, and GFP expression on alb1,afp, and ttr mRNA levels in H2.35, normalized to both actin and hprt. Results with mock-infected, undifferentiated samples were set to 1; n = 3. *p <0.05, **p <0.005 for designated conditions. (D) HepG2 were infected lentiviruses expressing Flag-Grg3 or Flag-Grg3-L36D/L85D mutant. Flag-tag (left) and albumin (right) immunofluorescences were merged in the right panels. Arrowheads denote cells expressing exogenous Flag-Grg3. All cells in the field are evident.
Next, we ectopically expressed Flag-tagged full length Grg3 from a lentiviral vector in HepG2, a human hepatoma cell line. The expression level of Grg3 was above that seen by endogenous Grg in the non-liver cell lines (Fig. 5A), considering that about 40% of the cells were infected (Fig. 5D). Notably, the subset of cells expressing Flag-Grg3 (green) expressed lower amounts of albumin protein (red) (Fig. 5D). We performed the same experiment using an N-terminally mutated form of Flag-tagged Grg3 that is deficient in transcriptional repression (Song et al., 2004), and found that this mutant did not suppress albumin expression (Fig. 5D). Taken together, above results indicate that Grg3 represses FoxA1 target genes and that the function is conserved between mouse and human cells.
FoxA1 recruits Grg3 to spread over a 3−4 nucleosome region in vivo
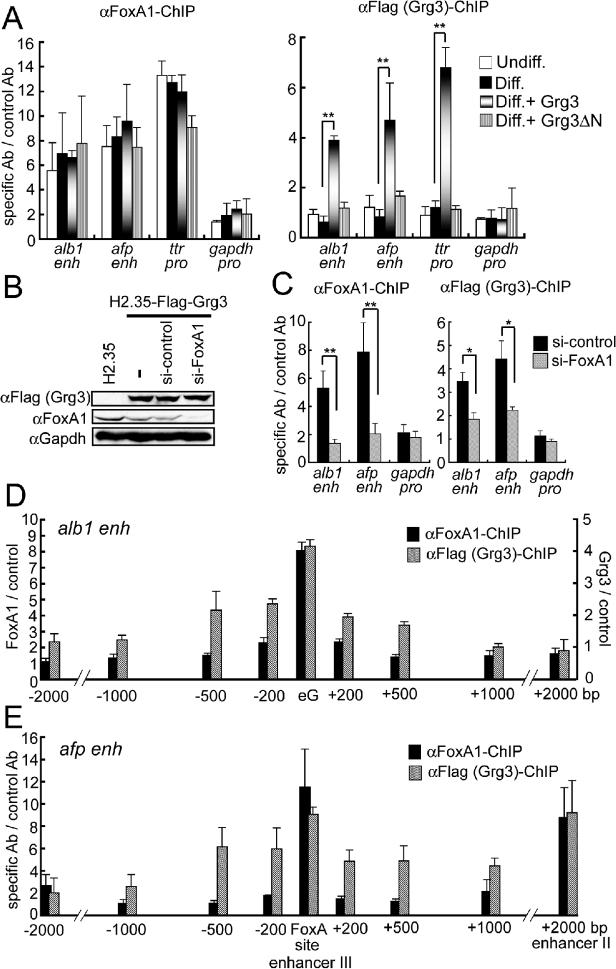
To assess the interaction of FoxA1 and Grg3 with alb1 and afp enhancer elements and the ttr promoter, which contain functional FoxA binding sites, we employed chromatin immunoprecipitation (ChIP) experiments on endogenous genes in H2.35 cells. We found that endogenous FoxA1 binds all three target elements in both differentiated and undifferentiated H2.35 cells, and infection with the Grg3 retroviruses had a minimal effect on FoxA1 binding (Fig. 6A, left). ChIP studies revealed that full-length Grg3 was bound to the DNA elements in infected cells, whereas Grg3-ΔN was not (Fig. 6A, right), consistent with the ability of only full-length Grg3 to inhibit transcription (Fig. 5C). To determine whether the binding of Grg3 to the FoxA targets depends upon FoxA1, we depleted endogenous FoxA1 by siRNA. The siRNA treatment decreased FoxA1 protein level significantly, while Gapdh and exogenous Flag-Grg3 levels were unchanged (Fig. 6B). We note that all cells expressing Flag-Grg3 exhibited somewhat lower levels of FoxA1, compared to uninfected cells (Fig. 6B), which we speculate is due to Grg3 weakly suppressing the endogenous FoxA1 gene. Regardless, only the siRNA specific to FoxA1 caused a marked decrease in FoxA1 binding the enhancers (Fig. 6C, left); ttr was not tested. Notably, Grg3 localization to both enhancers also decreased significantly (Fig. 6C, right). Since Grg3 localization was not reduced as much as that for FoxA1, other factors may be involved. Nonetheless, these results mimic the in vitro experiments in showing that FoxA1 can help recruit Grg3 to target sites in a genomic context.
Fig. 6. FoxA1 recruits Grg3 to blanket local enhancer regions.
(A) ChIP were performed on mock-infected undifferentiated H2.35, mock-infected, Flag-Grg3, and Flag-Grg3-ΔN retrovirus-infected, differentiated H2.35 cells with FoxA1 and Flag antibodies. The ratio between the scores obtained with the specific antibodies and control antibodies are shown with SD. (B) H2.35 expressing Flag-Grg3, with or without siRNA, were incubated for 96 hr at 37°C. Flag-Grg3, FoxA1, and Gapdh proteins were detected by Western blotting. (C) Depletion of FoxA1 decreased the binding of both FoxA1 and Grg3 to the alb1 and afp enhancers. H2.35 expressing Flag-Grg3 were transfected with control- or FoxA1-siRNA, incubated for 96 hr at 37°C, and subjected to ChIP. (D, E) FoxA1 localizes to its target sites, but Grg3 spreads locally from the FoxA1 recruitment site. ChIP were performed on Flag-Grg3-expressing, differentiated H2.35. ChIP products were analyzed with primer sets at the indicated distances from the FoxA1 sites at the alb1 enhancer (D) and afp enhancers III and II (E). n = 3. *p <0.05, *p <0.005.
Because the in vitro results implicated that Grg3 protein can form a resistant structure that extends several nucleosomes from the site of recruitment, we assessed the genomic range of Grg3 association around the FoxA1 site at the alb1 and afp enhancers in infected H2.35. By analyzing the ChIP products with primer sets extending several kb from the enhancers, we found that both FoxA1 and Grg3 bound most strongly to the FoxA1 target region, but only Grg3 exhibited reproducible binding that extended beyond the FoxA1 site (Fig. 6D). Specifically, while the FoxA1 ChIP products decreased to background levels by 200−500 bp on either side of the alb1 FoxA1 sites, the Grg3 ChIP products persisted to 40−50% of the maximum extending 500 bp from the FoxA1 sites. This compares favorably with the in vitro chromatin protection data elicited by FoxA1 recruitment of Grg3 in vitro (Fig. 4C). Similar results were obtained for the afp enhancer, where FoxA1 binding was specific to the target site, but Grg3 binding persisted for 500 bp upstream. Downstream of the afp enhancer, Grg3 persisted 1000 bp, but was accompanied by strong binding with FoxA1 at another enhancer 2000 bp downstream (Fig. 6E) (Millonig et al., 1995). We conclude that recruitment of Grg3 by FoxA1 to a single nucleosome is sufficient to blanket a domain that extends 3−4 nucleosomes, both in vitro and in vivo, about the size of a typical regulatory sequence. When bound to FoxA1 separated by ∼10 nucleosomes (2 kb), Grg3 appears to span the distance, but then diminishes distal to the FoxA sites.
Grg3-FoxA1 complexes in chromatin impair recruitment of other factors
We next assessed the mechanism by which Grg3 at FoxA1-bound chromatin impairs transcription at enhancers and promoters. During H2.35 cell differentiation, we found that NF-1, a transcriptional activator that is essential for the alb1 enhancer (Jackson et al., 1993), exhibits enhanced recruitment to the enhancer, while linker histone recruitment decreases slightly (Fig. 7B, black bars). We also found that TBP and RNA polymerase II, phosphorylated on the C-terminal repeat at serine 5, exhibit enhanced recruitment to the ttr promoter (Fig. 7B). Western blotting showed that the expression of Grg3 and Grg3-ΔN in H2.35 cells did not reduce the levels of NF-1, TBP, or linker histone (Fig. 7A). However, by ChIP, the increase in NF-1 occupancy at the alb1 enhancer in differentiated H2.35 was blocked by Grg3, but not by Grg3-ΔN (Fig. 7B). H1 occupancy was not significantly changed (Fig. 7B). At the ttr promoter, Grg3, but not Grg3-ΔN, blocked the increased occupancy of TBP and pol II/p-serine 5. Altogether, we conclude that, unlike other co-repressors, recruited Grg3 impairs the local recruitment of other factors that are crucial for enhancer and promoter activation.
Fig. 7. FoxA-recruited Grg3 impairs binding by transcriptional activators.
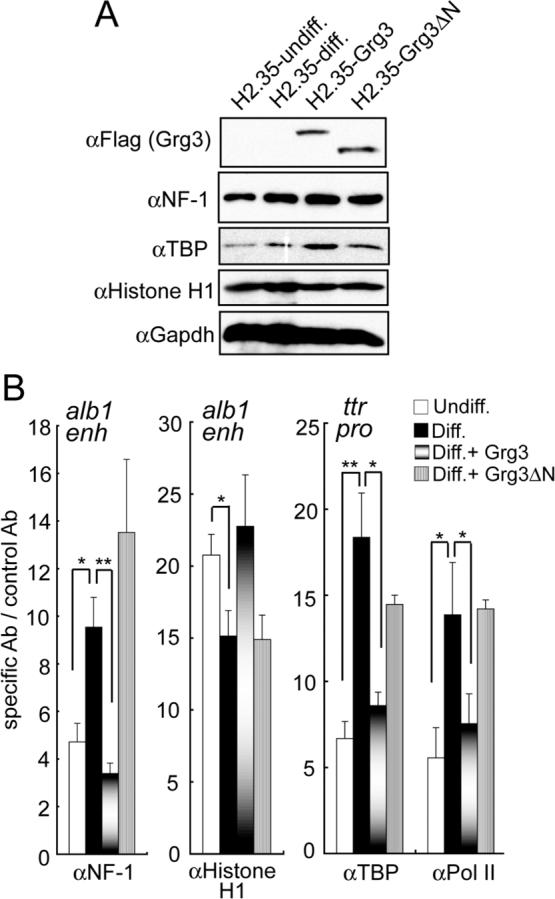
(A) Western blotting of whole cell lysates showing that ectopic expression of Grg3 isoforms does not reduce the levels of NF-1, TBP, or histone H1. (B) ChIP data (n=3) for the designated proteins at the alb1 enhancer or ttr promoter; pol II, anti-phosphorylated serine 5 of the C-terminal repeat. The data show that NF-1, TBP, and pol II exhibit enhanced recruitment to the designated regulatory elements during H2.35 cell differentiation (black bars) and that this recruitment is blocked by prior binding of Grg3 (graded bars); Grg3-deltaN had no effect (grey bars). *p <0.05, **p <0.005.
Discussion
Gro/TLE/Grg transcriptional corepressors regulate many developmental and signaling pathways, but their repressive mechanisms are not fully understood. We investigated Gro/TLE/Grg-mediated repression by using various chromatin substrates, both in vitro and in vivo, and present evidence for a newly discovered activity for this class of transcriptional corepressor.
In the absence of recruitment by a transcription factor, we found that Grg3 readily binds mononucleosome particles, at least in part via the interaction with the tails of histones H3 and H4. However, we discovered another level of Grg3 interaction with chromatin: with a dinucleosome subunit, helping to generate a condensed structure. In vivo studies suggest that the yeast Tup1 homolog bridges across adjacent nucleosomes (Ducker and Simpson, 2000). Since we found that N-terminally deleted Grg3, deficient in tetramerization (Chen et al., 1998), also elicits chromatin condensation, it is unlikely that condensation is mediated by bridging across nucleosomes by different Grg3 subunits in a native tetramer. We suggest that chromatin condensation may be generated by an individual Grg3 subunit interacting simultaneously with H3 tails of adjacent nucleosomes or with a new structure generated by the interface of adjacent nucleosomes. The ability of different domains of Tup1 to interact independently with core histones (Edmondson et al., 1996) is consistent with the former possibility.
By contrast, aggregation of separate nucleosome arrays requires an intact tetramerization domain of Grg3, as does the generation of recruitment-based, nuclease-resistant chromatin in vitro and transcriptional repression in vivo. Deletion of the tetramerization domain diminished, but did not eliminate, FoxA1 interactions, making it difficult to assess the extent to which the ability to aggregate arrays is necessary for the repressed chromatin structure upon FoxA1-based recruitment. Regardless, the middle region of Grg3 becomes exposed to V8 protease when full-length Grg3 binds to nucleosome arrays. Thus, chromatin binding induces a conformational change in the tetramer so that the subunits become exposed. This could facilitate the bridging of Grg3 between arrays to generate the aggregates. Secondarily, the conformational change in Grg3 induced by chromatin binding may indirectly facilitate stable interactions with FoxA, since we found that the middle region of free Grg3 impairs the protein from binding to FoxA factors.
Significantly, chromatin binding by Grg3 is indispensable for FoxA1- and Hes1-based genomic site recruitment; neither transcription factor could recruit Grg3 to free DNA. The existence of positioned nucleosomes at the alb1 enhancer in liver cells (McPherson et al., 1993; Chaya et al., 2001) may facilitate the ability of FoxA factors to recruit Grg3. In summary, we suggest that Gro/TLE/Grg proteins scan chromatin domains for being able to transiently bind nucleosomes, which stabilizes an open tetramer and consequently allows the tetramer to bind a recruiting transcription factor. Since Groucho/TLE/Grg proteins are thought to be refractory to binding highly acetylated chromatin (Chen and Courey, 2000), associated HDAC activity (Chen et al., 1999) could enable stable transcription factor-based recruitment.
Furthermore, we found that Grg3 generates a compact chromatin structure that is resistant to nucleases only when binding both chromatin and a recruiting transcription factor. We found that FoxA1-Grg3-array complexes are resistant to three different nucleases over the distance of approximately 3−4 nucleosomes in vitro. Strikingly, this corresponds to the same region over which Grg3 was recruited to the alb1 enhancer by FoxA1 in vivo, causing repression. Three to four nucleosomes spans the size of many enhancer and promoter elements. We therefore suggest that the recruitment-based blanketing of Grg3 over such a chromatin domain is part of the mechanism for transcriptional repression by the Gro/TLE/Grg corepressors.
Functionally, we found that recruitment-based “blanketing” of local chromatin by Grg3 results in impaired recruitment of a transcriptional activator, TBP, and RNA polymerase II. This distinguishes the mechanism of repression by Grg with that of Sir complexes (see Introduction). Strikingly, Grg3 impaired the enhanced recruitment of NF-1, TBP, and pol II that normally occurs when the H2.35 liver cell line is shifted to differentiating culture conditions. Interestingly, local levels of linker histone were not significantly affected by Grg3, indicating that the Grg3-bound chromatin neither excludes linker histone nor is excluded by it.
When Grg recruitment sites are separated in chromatin, as for the afp enhancers, Grgs can apparently span a larger chromatin domain; though this terminates distal to recruitment sites (Fig. 6E, −2000, −1000 regions). Although Gro/TLE/Grg proteins can repress genes distal to their site of recruitment (Chen and Courey, 2000), we suggest that by blanketing an enhancer, or an extended enhancer domain (as for afp), Gro/TLE/Grg proteins indirectly suppress distal enhancer-promoter interactions. This is opposed to long-range spreading of a repressive and resistant chromatin structure from a recruitment site, as seen with Sir proteins (Pirrotta and Gross, 2005). The yeast homolog Tup1 was found to spread along an entire STE6 gene on an episome (Ducker and Simpson, 2000). Interestingly, the linker regions of the episomal Tup1-STE6 chromatin remained sensitive to MNase, similar to the linker region sensitivity to nucleases of the condensed chromatin generated by Grg3 alone on our nucleosome arrays (Figs. 2E, 6A-D).
In conclusion, our studies emphasize the importance of reconstituting regulatory complexes on chromatin templates of different complexity and assessing the concordance with endogenous genes, to unveil mechanisms of genetic control. The mechanisms and consequences of chromatin binding that we found for Grg are different than those of other corepressor classes. Understanding how the condensed chromatin structure generated by Grg alone, in vitro, is converted to a resistant chromatin domain by transcription factor recruitment could unveil new molecular targets for antagonizing corepressor activity.
Experimental procedures
Quantitation
All graphs and plots showing quantitation were from experiments repeated at least three times. Error bars indicate standard deviations and p values are from Student's t test.
Protein purification, nucleosomal and DNA templates, binding reactions
The full length, ΔN, and middle Grg3 proteins were produced as His6-tagged forms using the Bac-to-Bac Baculovirus Expression System (Invitrogen). These and the ΔC and N-term Grg3, full-length FoxA1, FoxA2, Hes1, and ΔWRPW Hes1 proteins, were produced as His6-tagged forms in E.coli (Cirillo and Zaret, 1999). Grg3 reactions contained DTT to prevent nonspecific polymerization (Chen et al., 1998).
Albumin nucleosome arrays and EM analysis were as described (Cirillo et al., 2002), except that we used recombinant wild type and tailless histones (Luger et al., 1997; Dyer et al., 2004) and histone H1 was not present; Hes1 arrays are detailed in Sup. Fig. 2A. 208 bp sea urchin 5S rDNA probes for mononucleosome, or end-labeled 416 bp probes of two 5S genes for dinucleosomes were created by PCR with end-labeled primers. For a 33 bp DNA probe containing the eG FoxA1 binding site, the two oligo DNA; 5'-TTCCAAGGGAATGTTTGTTCTTAAATACCATCCC-3' and 5'-GGGATGGTATTTAAGAACAAACATTCCCTGGAA-3' were annealed and end-labeled.
Binding reactions for EMSA (Cirillo and Zaret, 1999; Cirillo et al., 2002) were loaded onto 4% polyacrylamide gels in 0.5X TBE buffer for DNA, mononucleosomes, and dinucleosomes, and 1% agarose gels in 0.5X TBE buffer for nucleosome arrays. Gels were exposed to X-ray film and a Fuji BAS 2500 phosphoimager, and analyzed with Multi Gauge software (Fuji).
Nuclease digestion assays
Binding reactions were carried out in 20 μl volumes. Diluted transcription factors were mixed with DNA or arrays in 10 mM HEPES pH 7.5, 0.25 mg/ml BSA, 2% glycerol, 5 mM DTT, 50 mM KCl. 35 or 70 ng of DNaseI diluted in 44 mM MgCl2 and 2.2 mM CaCl2 were added, followed by incubation at room temp for 1 min. EcoRI or XbaI digestions were carried out by adding 20 U enzyme (NEB) and incubating at 37°C for 2 min, upon which 20 μl of 20 mM Tris pH7.5, 50 mM EDTA, 1% SDS, 0.25 mg/ml tRNA, and 0.2 mg/ml proteinase K were added and incubation proceeded at 50°C for 1 hr. Purified products were analyzed on 1% agarose gels.
Cell culture, virus preparation and infection, and Western blotting
H2.35 were cultured as described (Zaret et al., 1988). HepG2 were cultured in Dulbecco's modified Eagle medium/Ham's F-12 medium 1:1, with 6% fetal bovine serum and 10 ug/ml insulin.
For retroviral vectors, Grg3 was PCR amplified from mouse embryo cDNA, Flag-tagged, and subcloned into pBabe-puro (Morgenstern and Land, 1990). pBabe-Flag-Grg3 or pBabe-GFP vectors were transfected into BOSC23 cells with FuGENE6 (Roche) and viral supernatants were collected after 48 h. Supernatants were mixed with culture medium at a ratio of 1:1, supplemented with 2 μg/ml polybrene, and added to H2.35 cells for 24 h. 72 hr later, cells were cultured for 4 days in medium with 2.5 μg/ml puromycin. For lentiviral vectors, Flag-tagged Grg3 was subcloned into pWPTS-GFP (with the GFP insert removed) (Zufferey et al., 1997). pWPTS-Flag-Grg3 was co-transfected with pCMV-Δ8.91 and pMDG (Zufferey et al., 1997) into 293T cells with FuGENE6 and viral supernatants were collected after 48 h. For infection, supernatants were mixed with culture medium at 1:1 and incubated with cells for 24 h.
Following infection, cells were lysed in 140 mM NaCl, 10 mM Tris pH 8.0, 1 mM EDTA, 1% Triton X-100, supplemented with Complete mini protease inhibitor cocktail (Roche). After removing debris by centrifugation, 20 μg protein samples were resolved by SDS-PAGE, transferred to PVDF membranes, and analyzed using anti-pan-Grg (Santa Cruz), GAPDH (RDI), GFP (Abcam), and Flag M2 (Sigma), followed by secondary antibodies conjugated with horseradish peroxidase and band detection by enhanced chemiluminescence.
RNA analysis
Total RNA was extracted using RNeasy Micro Kit (Qiagen), and reverse transcription was performed with iScript cDNA Synthesis Kit (Bio-Rad). PCR analysis was performed with an iCycler iQ multicolor real-time PCR detection system (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). All primer sets yielded a single product of the correct size. Relative expression levels were normalized to both actin and hprt.
Chromatin immunoprecipitation
Cells were crosslinked with 1% formaldehyde at room temp for 30 min, followed by adding glycine to 125 mM. Cells were washed three times in PBS, lysed, and genomic fragments were sonicated to a mean size of ∼400 bp. After removing insoluble material by centrifugation, ChIP was performed as described (Chaya et al., 2001). Antibody sources: NF-1 (Santa Cruz), H1 and TBP (Abcam), pol II anti-pS5 IgM (Covance), anti-IgM IgG (Sigma). Products were analyzed on an iCycler iQ multicolor real-time PCR detection system (Bio-Rad). PCR primer sets used are available upon request.
siRNA treatment
We used an siRNA pool against FoxA1 (siGENOME™ SMARTpool), and control siRNA (siCONTROL Non-targeting siRNA #1) from Dharmacon, transfecting with RNAiFect™ (Qiagen) by the manufacture's protocol for 96 hr at 37°C, then collecting samples for the ChIP and Western blotting.
Acknowledgements
We thank Michal Jarnik of the FCCC EM facility for electron microscopy, Isabel Cuesta for the tailless H3 and H4 plasmids, Didier Trono for lentivirus vectors, Hua-Ying Fan, Davide Ruggero, and Rugang Zhang for comments on the manuscript, and Eileen Pytko for help in its preparation. T.S. was supported by fellowships from the Japan Society for the Promotion of Science and the Uehara Memorial Foundation. The research was supported by grants to K.S.Z. from the NIH (R01 GM47903), the Mathers Charitable Foundation, and the NCI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Literature Cited
- Bossard P, Zaret KS. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel's Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, Zaret KS. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem. 2001;276:44385–44389. doi: 10.1074/jbc.M108214200. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol. 1998;18:7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L, Lin FR, Cuesta I, Jarnik M, Friedman D, Zaret K. Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Molecular Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- Costa RH, Grayson DR, Darnell JE., Jr. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Ducker CE, Simpson RT. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. Embo J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Saaib RD, Courey AJ. Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 2000;28:4189–4196. doi: 10.1093/nar/28.21.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Rowader KE, Stevens K, Jiang C, Milos P, Zaret KS. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol Cell Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lavigne M, Francis NJ, King IF, Kingston RE. Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, German MS. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- Liu JK, DiPersio CM, Zaret KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- McPherson CE, Shim E-Y, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely postioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Millonig JH, Emerson JA, Levorse JM, Tilghman SM. Molecular analysis of the distal enhancer of the mouse alpha-fetoprotein gene. Mol Cell Biol. 1995;15:3848–3856. doi: 10.1128/mcb.15.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acid Res. 1990;18:3587–3594. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr., Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol Cell. 2005;18:395–398. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Johnson AD. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- Song H, Hasson P, Paroush Z, Courey AJ. Groucho oligomerization is required for repression in vivo. Mol Cell Biol. 2004;24:4341–4350. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- Wang JC, Waltner-Law M, Yamada K, Osawa H, Stifani S, Granner DK. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3b. J Biol Chem. 2000;275:18418–18423. doi: 10.1074/jbc.M910211199. [DOI] [PubMed] [Google Scholar]
- Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 Physically Interact to Repress Transcription and Induce Mesoderm in Xenopus. J Biol Chem. 2007;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li P, Roeder RG, Wang Z. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol Cell Biol. 2001;21:4614–4625. doi: 10.1128/MCB.21.14.4614-4625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, DiPersio CM, Jackson DA, Montigny WJ, Weinstat DL. Conditional enhancement of liver-specific gene transcription. Proc Natl Acad Sci U S A. 1988;85:9076–9080. doi: 10.1073/pnas.85.23.9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.