Abstract
Three heterochromatin-like domains have been identified in S. cerevisiae that are refractory to transcription by Pol II, the silent mating-type loci, telomeres and the ribosomal DNA. Previous work has shown that chromatin remodelers can regulate silent chromatin. Here, we report the findings of an investigation into the role of ISW2 in transcriptional silencing at the rDNA. We show that the levels of retrotransposition and mRNA from a genetically marked Ty1 element located in the rDNA were increased significantly in isw2Δ cells, while transcript levels from Ty1 elements outside of the rDNA were not increased in cells lacking ISW2. Additionally, we show that Isw2 is not required for silencing at a telomere. Our findings demonstrate that Isw2 is required for transcriptional silencing at the rDNA and emphasize the differences in the regulation of transcriptional silencing at silent loci in S. cerevisiae.
Keywords: Ty1, retrotransposition, chromatin remodeler, gene silencing, telomeric silencing, heterochromatin
Introduction
In S. cerevisiae transcriptional silencing of Pol II-transcribed genes occurs at the homothallic mating (HM) loci, telomeres and the ribosomal DNA (rDNA) locus [1]. The rDNA locus on chromosome (chr) XII contains 150-200 copies of the ribosomal RNA (rRNA) genes. Each rDNA repeat has a Pol I-transcribed 35S rRNA gene and a non-transcribed spacer (NTS) (Fig. 1A). A number of studies have identified factors required for silencing at the rDNA including, but are not limited to, histones, histone-modifying enzymes and chromatin remodelers [2; 3; 4; 5; 6; 7; 8; 9; 10; 11; 12]. Interestingly, the requirement for these factors in silencing at the three silent loci in S. cerevisiae is not consistent. Set1, an enzyme that methylates lysine 4 of histone H3, is required for silencing at telomeres and the rDNA, but is not required for silencing of endogenous Pol II-transcribed genes at the HM loci [8; 13].
Fig. 1.
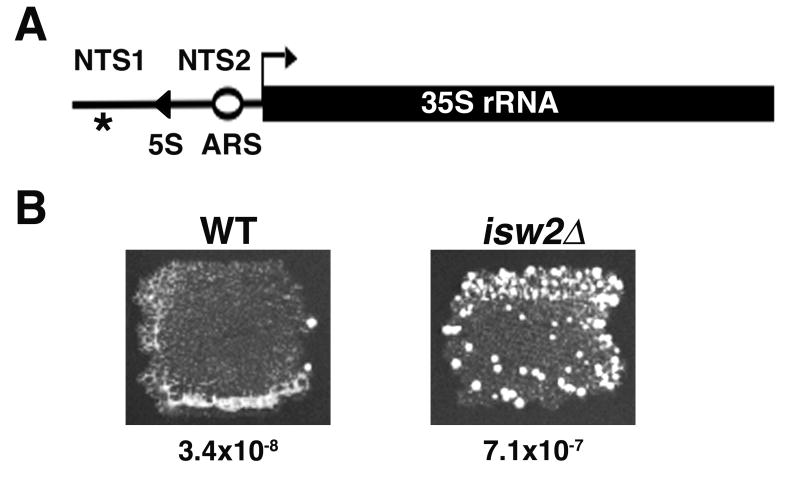
Deletion of ISW2 causes an increase in Ty1 retrotransposition. (A) A single rDNA repeat. Thick black line, 35S rRNA gene; thin line, NTS1 and NTS2; 5S rRNA gene, filled triangle; asterisk, location of Ty1his3AI-236; open circle, replication origin (ARS). (B) Colony growth and quantitative transposition assays measuring retrotransposition of Ty1his3AI-236 in wild-type and isw2Δ cells with rate of retrotransposition shown below each panel.
Chromatin remodeling proteins have been shown to regulate silencing at the rDNA. Deletion of SNF2 or SNF5, which encode members of the Swi/Snf complex, causes loss of silencing at the rDNA and telomeres [5]. The chromatin remodeling protein Isw1 is also required for silencing at the rDNA [7]. Isw1 is a member of the Imitation Switch (ISWI) family of chromatin remodelers [14]. Unlike members of the Swi/Snf complex, Isw1 functions in silencing at the HM loci but not at telomeres [7; 15]. Given that Isw1 associates with the rDNA and that deletion of ISW1 causes changes in rDNA chromatin structure, it has been proposed that Isw1 functions directly at the rDNA to regulate silencing [7].
A second ISWI family gene in S. cerevisiae, ISW2, is required for gene repression [16]. Cells lacking ISW2 overexpress a-specific genes in MATα cells and fail to repress early meiotic genes [17; 18]. Isw2 has been shown to repress individual genes as well, including INO1, PHO3 and CLB2 [19; 20; 21; 22]. Here we show that Isw2 is required for transcriptional silencing at the rDNA. Despite a strong rDNA-silencing defect, we did not detect changes in nucleosome positioning at the rDNA in isw2Δ cells. Additionally, we found that Isw2 is not required for silencing of a gene near a telomere. Consistent with previous work showing that Isw2 functions in gene repression, our data show that Isw2 is required for repression of Pol II transcription at the rDNA.
Materials and methods
Media and yeast strains
Media was prepared according to [23]. YPADT is YPD media supplemented with 20 mg/L L-tryptophan and 20 mg/L adenine sulfate. Synthetic complete (SC) and SC + 5-fluoroorotic acid (5-FOA) media were prepared as described [24]. Strains are listed in Table 1. MBY1198, MBY1912, MBY1914 and MBY1959 have been described previously [3; 7; 8]. ISW2 was replaced with the KANMX4 gene using PCR mediated gene disruption [25; 26]. The URA3 gene was integrated into the ADH4 locus near the telomere on the left arm of chr VII [27].
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| MBY1198 |
MATα ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0
Ty1his3AI-236 Ty1ade2AI-515 |
| MBY1699 | MBY1198 isw2Δ∷KANMX4 |
| MBY1912 | MBY1198 adh4∷URA3-TEL-VIIL |
| MBY1914 | MBY1198 set1Δ∷TRP1 adh4∷URA3-TEL-VIIL |
| MBY1959 | MBY1198 isw1Δ∷KANMX4 |
| MBY2148 | MBY1198 isw1Δ∷KANMX4 adh4∷URA3-TEL-VIIL |
| MBY2149 | MBY1198 isw2Δ∷KANMX4 adh4∷URA3-TEL-VIIL |
Ty1 retrotransposition
Cells were patched to YPADT agar plates, grown for 4 days at 20°C and printed to SC-His plates. Plates were photographed after 3 days at 30°C. Transposition rate experiments were performed as described [2]. Briefly, overnight cultures in YPADT liquid media were grown to saturation. Nine tubes containing 2 ml of YPADT were inoculated with ~500 cells from the overnight cultures and grown to saturation at 20°C. Three cultures were titered on YPADT plates to determine total cell number and cells from all nine cultures were spread on SC-His plates to determine the number of Ty1his3AI-236 retrotransposition events (His+). Transposition rates were calculated using the maximum likelihood method [28].
RNA Analysis
Northern blotting was performed as described [29]. Radiolabeled probes were used to detect Ty1his3AI-236, total Ty1, ACT1 RNA [8; 30]. Blots were quantified on a Molecular Dynamics Storm 860 phosphorimager using ImageQuant software.
Telomeric silencing assay
Spot growth assays to measure telomeric silencing were performed as described [8].
Micrococcal nuclease (MNase) accessibility
Spheroplasts were prepared and treated with MNase [31]. DNA was purified, ethanol precipitated, cleaved with PvuII and subjected to indirect end-labeling analysis using a radiolabeled probe specific for NTS2. Probe sequence is available upon request.
Results and Discussion
Deletion of the ISW2 gene causes increased Ty1 retrotransposition
We use an rDNA silencing system that takes advantage of Ty1 elements, naturally occurring retrotransposons in S. cerevisiae that are transcribed by Pol II. The Ty1 retrotransposition cycle is similar to the life cycle of retroviruses. Within a yeast cell, mRNA from Ty1 elements is packaged within a virus-like particle where it serves as a template for synthesis of cDNA by the Ty1-encoded reverse transcriptase. The resulting Ty1 cDNA is integrated back into the nuclear genome of the same cell completing the Ty1 retrotransposition cycle. A genetically marked Ty1 element, Ty1his3AI-236, used here is present in one of the rDNA repeats on chr XII. In wild-type cells, Ty1his3AI-236 is transcriptionally silenced and exhibits a low rate of transposition [2]. When rDNA silencing is disrupted, Ty1his3AI-236 is transcribed by Pol II leading to a higher rate of retrotransposition. Ty1his3AI-236 contains a modified his3 allele that, after retrotransposition, can be converted to a functional HIS3 gene [32]. Simple plate assays and quantitative experiments measuring His+ colony formation provide an indication of the rate of retrotransposition of the genetically marked Ty1 element.
The requirement for chromatin remodeling proteins in transcriptional silencing at the rDNA [5; 7] prompted us to investigate whether Isw2 plays a role in rDNA silencing. First, we measured transposition of Ty1his3AI-236 located in the rDNA (Fig. 1A) in wild-type and isw2Δ cells using a simple patch assay. We found that wild-type cells had a relatively low level of Ty1his3AI-236 transposition based on the small number of His+ colonies that grew on SC-His media (Fig. 1B). This low level of retrotransposition reflects transcriptional silencing of Ty1his3AI-236 in the rDNA [2]. In contrast, in isw2Δ cells, a greater number of His+ colonies grew on SC-His (Fig. 1B) indicating a high level of retrotransposition of the Ty1his3AI-236 element.
We performed quantitative measurements of the rate of retrotransposition of the Ty1his3AI-236 element located in the rDNA. Measurement of the rate of transposition (transposition events/cell/generation) for Ty1his3AI-236 is calculated from the number of His+ prototrophs generated after growth in non-selective rich media (Materials and methods). The rate of transposition of the Ty1his3AI-236 element in wild-type cells was 3.4 × 10-8/cell/generation and in isw2Δ cells was 7.1 × 10-7/cell/generation, indicating that the rate of transposition of the Ty1his3AI element was increased 20.8-fold in cells lacking ISW2 (Fig. 1B).
Isw2 regulates transcriptional silencing at the rDNA
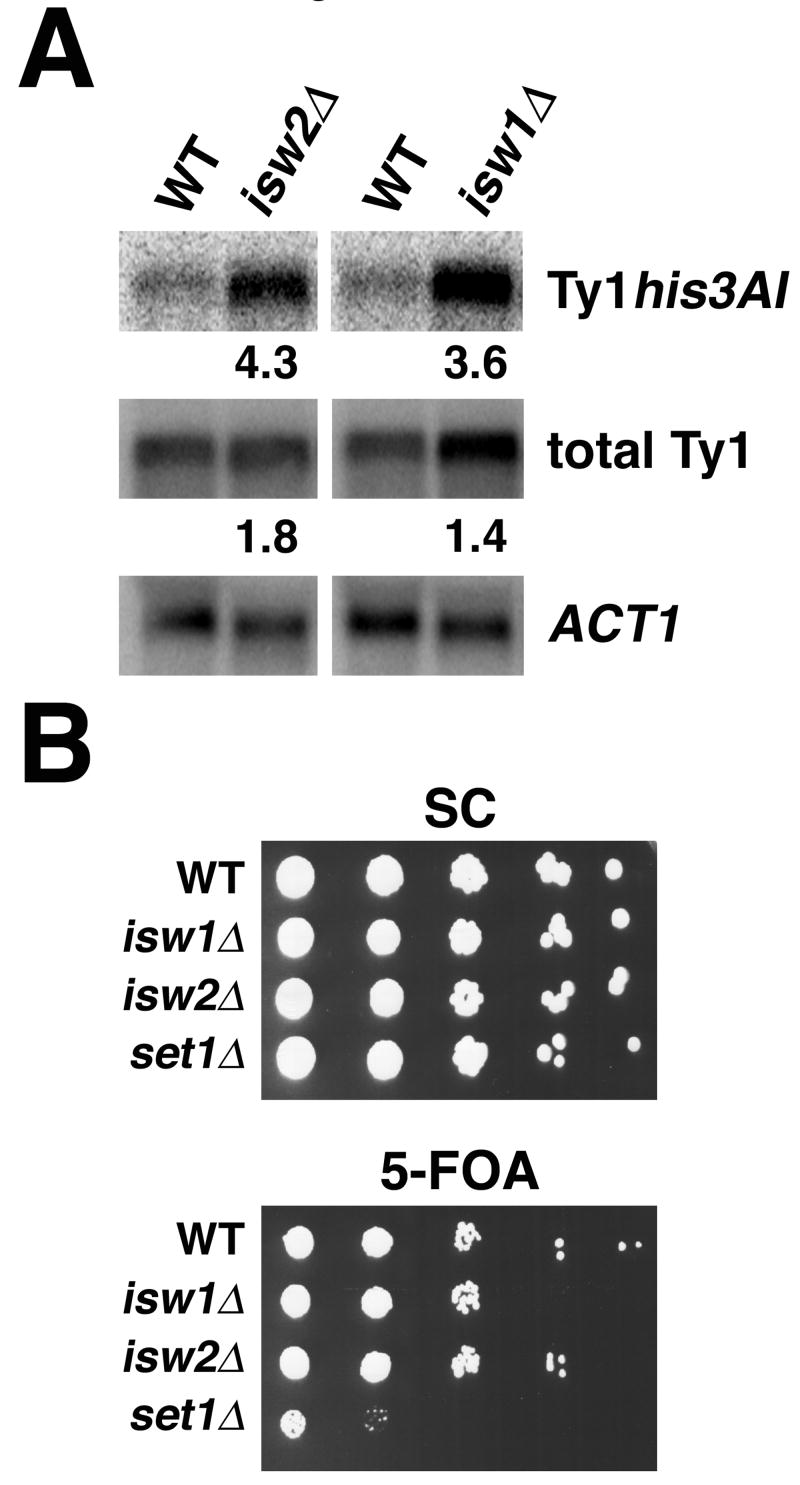
To determine if the higher rate of Ty1his3AI-236 transposition observed in isw2Δ cells was due to an increased level of Ty1his3AI mRNA, we measured steady-state Ty1his3AI transcript levels in wild-type and isw2Δ cells. Ty1his3AI-236 mRNA was detected by hybridization with a probe specific to his3 sequences (Fig. 2A, upper panel). The low level of Ty1his3AI mRNA observed in wild-type cells is consistent with transcriptional silencing at the rDNA. In isw2Δ cells, Ty1his3AI mRNA levels were increased 4.3-fold relative to the level in wild-type cells, indicating that Isw2 regulates transcriptional silencing at the rDNA. As a control, we performed Northern analyses on RNA from cells lacking ISW1, a chromatin remodeling protein that is closely related to Isw2 and is required for silencing at the rDNA and HM loci [7; 15]. As expected, Ty1his3AI RNA levels were increased in isw1Δ cells.
Fig. 2.
Isw2 is required for transcriptional silencing at the rDNA. (A) Total RNA from wild-type, isw2Δ and isw1Δ cells were analyzed to determine the levels of Ty1his3AI-236 mRNA (upper panel), total Ty1 mRNA (middle panel) and ACT1 mRNA (lower panel). Transcript levels for Ty1his3AI-236 and total Ty1 were normalized to levels of ACT1 or PYK1 (not shown) mRNA. The average ratios of normalized Ty1his3AI-236 mRNA in mutants relative to that in wild-type cells are shown below each panel (n=3). These values +/-SE were: isw2Δ, 4.3+/-0.7, p=0.011; isw1Δ, 3.6+/-0.8, p=0.036. The average ratios of normalized total Ty1 transcript in mutants relative to wild-type cells are shown below each panel (n=3). These values +/-SE were: isw2Δ, 1.8+/-0.6, p=0.24; isw1Δ, 1.4+/-0.2, p=0.12. (B) Wild-type, isw1Δ, isw2Δ and set1Δ cells were analyzed to evaluate silencing of a URA3 reporter gene at a telomere. Five microliters of 10-fold serial dilutions of stationary phase cultures were spotted to SC (upper) and SC+5FOA (lower) media.
Approximately 30 endogenous Ty1 elements have been mapped in the S. cerevisiae genome and all of these are located outside of the rDNA. To determine if deletion of ISW2 causes a general increase in transcription of endogenous Ty1 elements, we measured total Ty1 mRNA (Fig. 2A, middle panel). Consistent with previous studies [20; 33], no significant difference in steady-state total Ty1 mRNA levels were observed in wild-type and isw2Δ cells. In contrast to the regulation of the Ty1his3AI-236 element in the rDNA, cells lacking either ISW1 or ISW2 individually do not exhibit defects in transcription of genomic Ty1 elements [7; 20]. However, previous work has shown that cells lacking both ISW1 and ISW2 have higher levels of Ty1 transcripts than wild-type or single ISW1 or ISW2 deletion mutants, a result that suggests that Isw1 and Isw2 act redundantly to repress transcription of genomic Ty1 elements [20]. Consistent with transcriptional silencing of Pol II genes at the rDNA being a result of specialized silent chromatin, data presented here and elsewhere [7] suggest that Isw1 and Isw2 alter the function of silent chromatin at the rDNA. However, in contrast to the regulation of endogenous Ty1 elements located outside of the rDNA [20], our results suggest that ISW1 and ISW2 do not act redundantly in the silencing of Pol II-transcribed genes in the rDNA.
There is precedence for the regulation of the rDNA locus by the ISWI family chromatin remodelers. In S. cerevisiae, Isw1 associates with the rDNA and is required for the silencing of Pol II-transcribed genes at the rDNA [7]. In mammals, Snfh2, an ISWI chromatin remodeling protein and member of the nucleolar remodeling complex NoRC, is required for silencing of Pol I transcription [34; 35]. Here, we show that Isw2 plays a role in the regulation of Pol II transcription at the rDNA in S. cerevisiae, however, the lack of an effect of deletion of ISW2 on total rRNA levels or cell-growth properties (data not shown) suggests that Isw2 does not regulate transcription of the ribosomal RNA genes by Pol I in S. cerevisiae.
Isw2 is not required for transcriptional silencing at telomeres
Previous work has shown that deletion of ISW2 does not significantly affect transcriptional silencing near a telomere [36]. These studies were performed in S. cerevisiae strains of the W303 genetic background. Differences in some phenotypes exhibited by isw2Δ cells have been observed upon comparison of W303 and S288C strains, where variations in the degree of derepression of a-specific genes in MATα cells has been reported [37]. To address the possibility that Isw2 functions differently in transcriptional silencing in W303 vs S288C strains, we analyzed telomeric silencing in S288C strains that lack ISW2. We performed a plate growth assay with cells that contain a URA3 reporter gene at the telomere on the left arm of chromosome VII. The URA3 gene product converts 5-FOA to 5-fluorouracil, which is toxic to cells. In wild-type cells, where the URA3 gene is silenced, robust growth was observed on media containing 5-FOA (Fig. 2B, lower panel). Growth of isw2Δ cells was similar to that of wild-type cells, indicating that Isw2 is not required for telomeric silencing in the S288C strain background. Set1 has been shown to be required for telomeric silencing [8; 13]. We found that growth of set1Δ cells on SC+5-FOA plates was greatly reduced, reflecting a loss of telomeric silencing and expression of the URA3 gene (Fig. 2B). As a negative control, we assayed isw1Δ cells that do not exhibit defects in telomeric silencing [15]. In summary, our data show that in S288C strains Isw2 is not required for telomeric silencing, similar to what has been reported for W303 strains [36].
Deletion of Isw2 does not alter nucleosome positioning at the rDNA
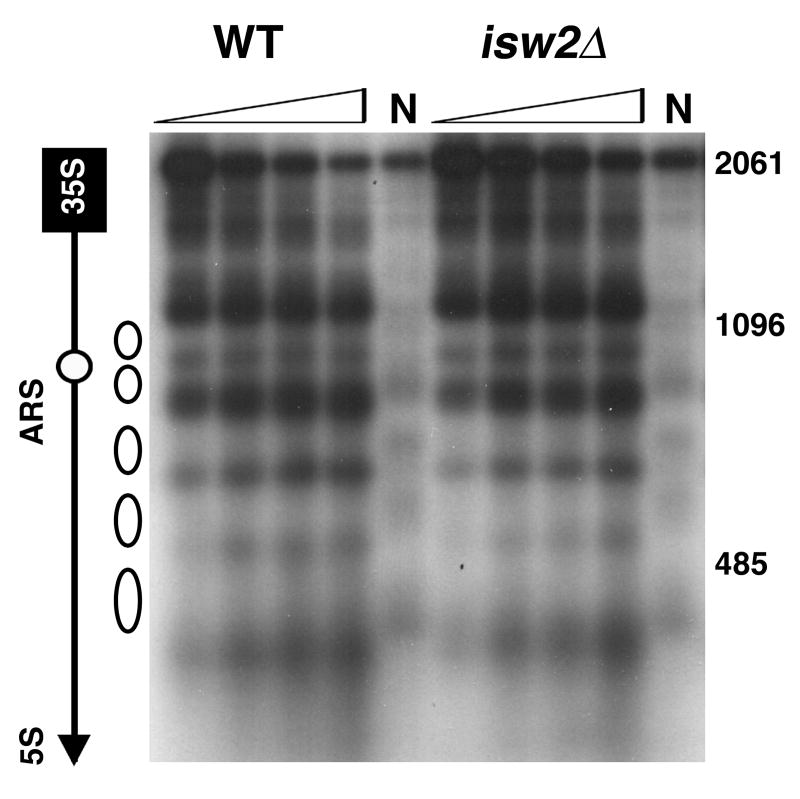
We performed MNase accessibility studies to determine if Isw2 regulates chromatin structure at the rDNA like other rDNA silencing factors [6; 7; 38]. Our data show that nucleosome positioning at the NTS2 region of the rDNA where positioned nucleosomes have been identified [38; 39] is similar in wild-type and isw2Δ cells (Fig. 3). We also examined chromatin structure of NTS1 and the 5S rRNA gene and found no differences in MNase accessibility patterns in wild-type and isw2sΔ cells (data not shown). Thus, while Isw2 is required for transcriptional silencing at the rDNA, isw2Δ cells do not exhibit detectable changes in chromatin structure at this locus.
Fig. 3.
Deletion of ISW2 does not alter nucleosome positioning at the rDNA NTS. Open triangles, increasing amounts of MNase; N, naked DNA; open ovals, five nucleosomes previously mapped to NTS2 [38; 39]. DNA marker (bp), on right. Other labels, as in Fig. 1A.
In this study we demonstrate a requirement for Isw2 in transcriptional silencing at the rDNA. While Isw2 is required for rDNA silencing, it is not required for silencing at telomeres. Indeed, these findings draw attention to differences in the regulation of heterochromatin-like domains in S. cerevisiae. Because deletion of ISW2 does not affect chromatin structure at the rDNA, we cannot determine if rDNA silencing requires direct association of the Isw2 protein with the rDNA. Interestingly, recruitment of Isw2 to sequences upstream of tRNA genes requires Bdp1, a subunit of the Pol III transcription factor TFIIIB [40]. Thus, it is not unreasonable to speculate that Bdp1 recruits Isw2 to the Pol III-transcribed 5S rRNA gene in the rDNA, an association that may contribute to Isw2’s role in rDNA silencing. Future studies examining the physical and genetic interactions of Isw2 with the rDNA and other silencing factors will provide greater insight into the role of Isw2 in rDNA silencing.
Acknowledgments
We are grateful to Trisha S. Green for initial input. This work was supported by grant GM-070930 from the National Institutes of Health to MB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 2.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–69. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 3.Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol. 2002;12:165–70. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- 4.Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–57. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dror V, Winston F. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:8227–35. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller JE, Bryk M. Isw1 Acts Independently of the Isw1a and Isw1b Complexes in Regulating Transcriptional Silencing at the Ribosomal DNA Locus in Saccharomyces cerevisiae. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–67. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy N, Runge KW. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr Biol. 2000;10:111–4. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- 10.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–54. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 11.Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–97. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–56. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 13.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 14.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–47. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 15.Cuperus G, Shore D. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics. 2002;162:633–45. doi: 10.1093/genetics/162.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellor J, Morillon A. ISWI complexes in Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1677:100–12. doi: 10.1016/j.bbaexp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–33. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz C, Escribano V, Morgado E, Molina M, Mazon MJ. Cell-type-dependent repression of yeast a-specific genes requires Itc1p, a subunit of the Isw2p-Itc1p chromatin remodelling complex. Microbiology. 2003;149:341–51. doi: 10.1099/mic.0.25920-0. [DOI] [PubMed] [Google Scholar]
- 19.Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–60. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–26. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherriff JA, Kent NA, Mellor J. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol Cell Biol. 2007;27:2848–60. doi: 10.1128/MCB.01798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugiyama M, Nikawa J. The Saccharomyces cerevisiae Isw2p-Itc1p complex represses INO1 expression and maintains cell morphology. J Bacteriol. 2001;183:4985–93. doi: 10.1128/JB.183.17.4985-4993.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose MD, Winston F, Hieter P. Methods in Yeast genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 1990. [Google Scholar]
- 24.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–75. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 25.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucl Acids Res. 1993;21:3329–30. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–87. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 28.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 29.Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–19. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curcio MJ, Garfinkel DJ. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol. 1992;12:2813–25. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–7. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curcio MJ, Garfinkel DJ. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A. 1991;88:936–40. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–54. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Langst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–41. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strohner R, Nemeth A, Nightingale KP, Grummt I, Becker PB, Langst G. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol. 2004;24:1791–8. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iida T, Araki H. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:217–27. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trachtulcova P, Frydlova I, Janatova I, Dorosh A, Hasek J. The W303 genetic background affects the isw2 delta mutant phenotype in Saccharomyces cerevisiae. Folia Microbiol (Praha) 2003;48:745–53. doi: 10.1007/BF02931508. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Mueller JE, Bryk M. Sir2 Represses Endogenous Polymerase II Transcription Units in the Ribosomal DNA Nontranscribed Spacer. Mol Biol Cell. 2006;17:3848–59. doi: 10.1091/mbc.E06-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelauer M, Cioci F, Camilloni G. DNA protein-interactions at the Saccharomyces cerevisiae 35 S rRNA promoter and in its surrounding region. J Mol Biol. 1998;275:197–209. doi: 10.1006/jmbi.1997.1451. [DOI] [PubMed] [Google Scholar]
- 40.Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 2005;19:955–64. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]