Summary
The 20S proteasome functions in protein degradation in eukaryotes together with the 19S ATPases or in archaea with the homologous PAN ATPase complex. PAN and the 19S ATPases contain a conserved C-terminal hydrophobic-tyrosine-X motif (HbYX). We show that these residues are essential for PAN to associate with the 20S and open its gated-channel for substrate entry. Upon ATP binding, these C-terminal residues bind to pockets between the 20S’s α-subunits. Furthermore, seven-residue peptides from PAN’s C-terminus that contain the HbYX motif also bind to these sites and induce gate-opening in both archaeal and mammalian 20S proteasomes. Gate-opening could be induced by short C-terminal peptides from the 19S ATPase subunits, Rpt2 and Rpt5, but not by ones from PA28/26, which lack the HbYX motif and cause gate-opening by distinct mechanism. The C-terminal YX residues in the 19S ATPases were also shown to be critical for gating and stability of mammalian and yeast 26S proteasomes. Thus, the C-termini of the proteasomal ATPases function like a “key-in-a-lock” to induce gate-opening and allow substrate entry.
Introduction
The 26S proteasome is a large ATP-dependent proteolytic complex that catalyzes the degradation of most cellular proteins (Glickman and Ciechanover, 2002; Voges et al., 1999), generally after linkage to a ubiquitin chain (Glickman and Ciechanover, 2002; Goldberg, 2005). Proteasomes are also found in archaea and certain eubacteria, where ubiquitin is not present, but proteolysis is still ATP-dependent (Goldberg, 2005). The active forms of proteasomes in archaea and eukaryotes are composed of an ATPase complex bound to one or both ends of the 20S core proteasome (DeMartino and Slaughter, 1999; Voges et al., 1999). This hollow cylinder is composed of four stacked rings (Groll et al., 1997; Lowe et al., 1995). Its outer rings contain seven α-subunits that surround a narrow channel, through which polypeptides enter the 20S’s central chamber, where its multiple proteolytic sites are located. The passage of proteins and peptides through this channel is restricted by the N-termini of the α-subunits, which function as a gate (Groll et al., 2000; Groll and Huber, 2003). The ATPase ring complexes open this gate and thus regulate proteolysis by the 20S particle (Benaroudj et al., 2003; Kohler et al., 2001; Smith et al., 2005). In eukaryotes, the base of the 19S regulatory complex contains six different ATPases (Rpt1–6) (Fu et al., 2001).
In archaea, the proteasome-regulatory complex, PAN, is composed of six identical subunits that are close homologs of the 26S ATPases (Benaroudj et al., 2003; Navon and Goldberg, 2001; Ogura and Tanaka, 2003; Zwickl et al., 1999). In electron micrographs, PAN resembles a top-hat structure capping the 20S cylinder and appears similar to densities in the base of the 19S complex (Smith et al., 2005). These ATPase rings associate with the outer α-ring of the 20S, where in the presence of ATP, they unfold globular proteins and promote their translocation through the gated entry channel into the 20S particle (Benaroudj et al., 2005; Kohler et al., 2001; Smith et al., 2005; Voges et al., 1999).
The simpler PAN-20S complex offers major advantages for exploring proteasome function (Benaroudj et al., 2003; Navon and Goldberg, 2001; Ogura and Tanaka, 2003; Smith et al., 2005). For example, it does not require ubiquitin conjugation for proteolysis, the archaeal 20S contains only one type of α and β subunits, and these complexes assemble when expressed in E. coli. Our recent studies have established that upon binding of ATP or nonhydrolyzable ATP analogs, PAN associates with the 20S particle and triggers gate opening in the α-ring. This gate-opening by PAN, even without ATP hydrolysis, facilitates the diffusion of peptides and unfolded proteins into the 20S. Similarly, ATP binding in the eukaryotic 26S complex also causes gate opening and entry of unfolded proteins into the 20S (Smith et al., 2005).
The present studies were undertaken to investigate the mechanism by which these ATPases induce gate opening in the 20S proteasome. One attractive model for gate opening comes from studies by Hill and colleagues on a very different type of proteasomal regulator, PA26, the 11S invertebrate homolog of the mammalian PA28 γ complex and the PA28αβ complex that stimulates antigen presentation (Goldberg et al., 2002). Though the biological functions of PA26 and PA28 γ are not clear, these heptametric complexes associate with the ends of the 20S particle and facilitate entry of short peptides (DeMartino and Slaughter, 1999). The association of PA28/26 with the 20S requires their extreme C-termini (Ma et al., 1993), and by X-ray crystallography, Hill and co-workers demonstrated that these C-termini dock into pockets between adjacent α-subunits, providing binding energy for the PA26-20S complex (Forster et al., 2005; Whitby et al., 2000). However, this binding of PA26’s C-termini to the α-intersubunit pockets is not sufficient to induce 20S gate opening (Zhang et al., 1998), which instead requires an additional domain distant from its C-termini. This “activation domain” is presented by PA26 in a 7-fold symmetric circular array that interacts with the bases of the N-terminal gating residues of the 7 α-subunits. This interaction destabilizes the asymmetrical closed gate conformation of the 20S proteasome and stabilizes its symmetric open-gated conformation (Forster et al., 2003).
While it is not known how the proteasomal ATPases induce gate opening, two groups have suggested that these ATPases use a mechanism like that of PA26 (Forster et al., 2005; Kohler et al., 2001), which requires an “activation-loop”. However, PA28/26 and the proteasomal ATPases are very different protein complexes and do not share any sequence homology. While PA28/26 are heptameric rings, like the 20S proteasome with which they form a matched 7:7 subunit interface, the proteasome-regulatory ATPases, PAN, and those in the base of the 19S (Rpt1–6) are hexameric and thus form an asymmetrical complex with the 20S with a mismatched 6:7 subunit interface. Since PA26 applies its symmetry to the 20S α-ring to induce gate opening (Forster et al., 2005; Forster et al., 2003), the asymmetrical proteasomal ATPase complexes should not be able to induce gate opening in the same fashion. Furthermore, PA28/26 appear to be static complexes, whereas the proteasomal ATPases, like other AAA ATPases, are highly dynamic structures that undergo large conformational changes upon binding and hydrolysis of ATP (DeLaBarre and Brunger, 2005; Wang et al., 2001). These major differences suggest that the proteasomal ATPases and PA28/26 probably utilize different mechanisms to stabilize the open-gate conformation of the α-subunits N-termini.
To investigate how proteasomal ATPases induce gate opening, we first tested whether PAN’s C-termini are important for its ATP-dependent association with the 20S and its ability to induce gate opening in the 20S proteasome. We show here 1) that PAN’s C-termini contain a conserved short domain which binds to the 20S and acts like a “key-in-a-lock” to cause gate opening, and 2) that a similar mechanism functions in gating in the more complex eukaryotic 26S proteasome.
Results
The proteasomal ATPases’ C-termini contain a conserved HbYX motif
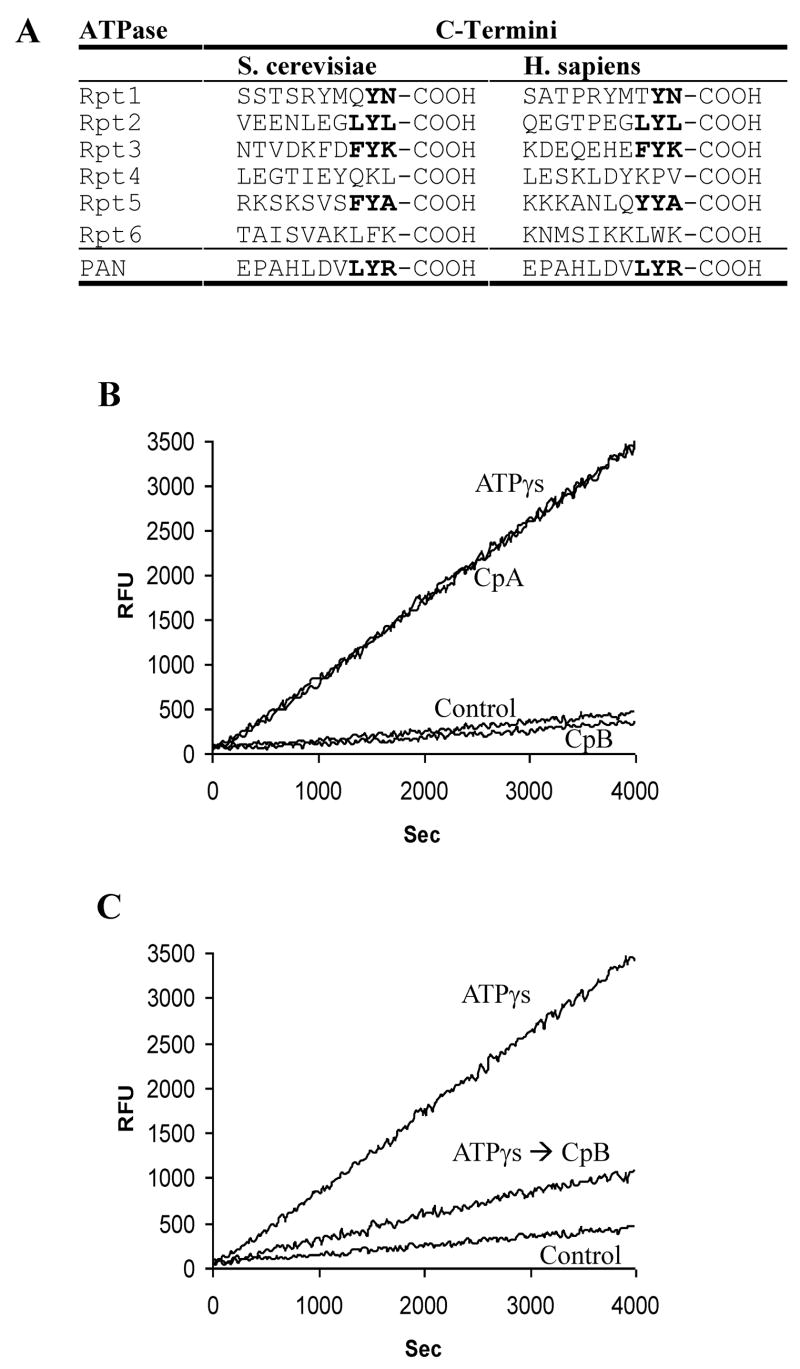
If the extreme C-termini of these ATPases are important for its association with the 20S, then the critical residues may be conserved, since the requirements for complex formation and gate opening by PAN and the 19S ATPases appear similar (Smith et al., 2005). We therefore compared their C-terminal sequences in various archaea and eukaryotes (Fig. 1A). While the terminal arginine residue in PAN is not conserved, the penultimate residue is a tyrosine in PAN from 9 different archaeal species (see Supplement Table S1), although in five archaeal species, it is a phenylalanine. A penultimate tyrosine is also found in the four ATPases, Rpt1, Rpt2, Rpt3 and Rpt5 (but not Rpt4 and Rpt6) in 26S proteasomes from humans, rats, mice, drosophila, arabidopsis, nematodes (except Rpt1) and yeast. A hydrophobic residue precedes the penultimate tyrosine in PAN from all 14 archaea species and also in Rpt2, Rpt3, Rpt5, and Rpt6 of these eukaryotes. Thus the conserved C-terminal motif, HbYX, where Hb is a hydrophobic residue, Y is a tyrosine (or in some archaea, a phenylalanine), and X can vary widely, appears in nearly all known proteasomal ATPases. Interestingly, no such sequence is found in the C-termini of PA26 and PA28α, β or γ.
Figure 1.
A) Carboxypeptidase treatment of the proteasomal ATPase regulatory complexes’ eliminates their ability to stimulate the 20S for peptide hydrolysis. A) Two residues preceding the C-terminal arginine in PAN are conserved in the eukaryotic 19S proteasome-regulatory ATPases. B) A solution of PAN, 20S and the fluorescent peptide LFP (Mca-AKVYPYPME-Dpa-amide) was pre-incubated with Carboxypeptidase A (CpA), B (CpB) or without either carboxypeptidase (control and ATPγS) for 5 minutes followed by addition of a general Carboxypeptidase inhibitor from potato tuber, (0.01mg/ml). H2O (Control), or ATPγS (ATPγS, CpA, CpB) was added, and the rate of LFP hydrolysis monitored in real-time. Both CpA and CpB were used at a final concentration of 0.08 Units/ml. C) Same as in A except ATPγS was added prior to addition of CpB. Values are mean’s of three independent experiments, error bars indicate standard deviations and all of these experiment were performed at least three times with similar results.
PAN’s C-terminal residues are required to stimulate 20S gate opening
To determine if PAN requires its C-terminal arginine to associate with the 20S proteasome, PAN and the 20S were pretreated with carboxypeptidase B (CpB), which specifically removes basic C-terminal residues, and after addition of ATPγS, complex formation and gate opening in the 20S were assayed (Smith et al., 2005). The stimulation of gate opening was monitored by measuring the hydrolysis of the quenched 9-residue fluorogenic substrate, LFP, whose entry into the 20S is very slow when the gate is closed (Smith et al., 2005). Treatment with carboxypeptidase B did not affect the ability of the 20S proteasome to hydrolyze LFP and did not alter PAN’s ATPase activity or its ability to unfold GFP-ssrA (Supplement Fig. S1 a and b). However, after carboxypeptidase B treatment, which should remove the arginine but not the penultimate tyrosine, PAN completely lost its ability to stimulate peptide entry (Fig. 1C). By contrast, carboxypeptidase A, which cleaves primarily after hydrophobic residues, had no effect on the stimulation of peptide entry. Thus, PAN’s C-terminal arginine is required for gate opening, which can account for the recent finding that this residue is also required for the stimulation of casein degradation by PAN (Forster et al., 2005).
Interestingly, if the PAN-20S complex was formed first by addition of ATPγS before the treatment with carboxypeptidase B, then PAN retained some ability to stimulate peptide hydrolysis (Fig. 1C). Therefore, PAN’s C-terminal residues are essential for activation of gate opening and are accessible to carboxypeptidases when the ATPase complex is not associated with the 20S, but become less accessible once the ATPase-20S complex is formed. (It is noteworthy that this protection from carboxypeptidase inactivation is only partial, probably because the association between PAN and the 20S is transitory (Smith et al., 2005). Thus, the C-terminus is likely to be located in the interface of the ATPase-20S complex, as shown below.
To investigate further the importance of PAN’s C-terminal arginine in gate-opening, we performed site-directed mutagenesis to remove it, replace it with other amino acids, or extend the C-terminus. When this arginine was deleted, PAN lost its ability to stimulate gate opening in the presence of ATPγS (as was found after treatment with carboxypeptidase B). However, mutating this arginine (R430) to alanine or tryptophan did not reduce PAN’s ability to stimulate the 20S, and mutation to a glycine (R430G) decreased it only slightly (Table 1). Because the R430G mutation still permitted gate opening, a side chain on the ultimate residue is not essential for 20S activation. Although many amino acids can replace this terminal arginine, surprisingly, replacement by a leucine markedly decreased PAN’s ability to stimulate the 20S, and replacement by an aspartate completely prevented activation. Furthermore, addition of an alanine C-terminal to the arginine blocked PAN’s ability to stimulate gate opening, even though an alanine in place of the arginine did not influence this activity. Thus, the binding site for the C-terminal sequences has some specificity, and while the ultimate residue does not require any specific side chain, basic side chains in this position do not allow function.
Table 1.
Mutagenesis to remove PAN’s C–terminal arginine destroys its ability to stimulate gate opening, but its replacement by certain other residues allows wild type activity
| PAN Sequence | Peptide Hydrolysis (% WT) |
|---|---|
| AHLDVLYR430 (WT) | 100 |
| AHLDVLY- | 4 |
| AHLDVLYD | 5 |
| AHLDVLYA | 100 |
| AHLDVLYW | 106 |
| AHLDVLYL | 13 |
| AHLDVLYG | 77 |
| AHLDVLYRA | 2 |
| AAAAAHLDVLYR | 74 |
| No PAN | 5 |
PANs C-terminal residue was mutated and the indicated PAN mutant (1ug) was incubated with 20S (0.25ug) in a 100ml reaction buffer and the rate of LFP hydrolysis was monitored in the presence or absence of ATPgS (20mM). Shown is the mean values of 3 or more experiments were the +/−SD was less than 5%.
The penultimate HbY residues are essential for PAN to stimulate gate opening
To determine if the conserved hydrophobic and tyrosine residues in the HbYX motif are important for the association with the 20S and stimulation of gate opening, we systematically mutated these residues. Tyrosine Y429 was found to be absolutely essential for PAN’s activity (Table 2). When it was mutated to any of eight other residues, including hydrophobic, aromatic, or charged, none stimulated LFP hydrolysis. This lack of stimulation with a phenylalanine in this position was surprising since the penultimate residue in PAN from five archaeal species is a phenylalanine. Mutating the leucine preceding the tyrosine to any of 10 other amino acids confirmed that only hydrophobic (Hb) residues supported activity, but replacement by an arginine, aspartate, cysteine or proline prevented the stimulation of gate opening.
Table 2.
The penultimate Tyr-429 and preceding hydrophobic residue Leu-428 in PAN are required for 20S gate opening
| Peptide Hydrolysis (% WT)
| ||
|---|---|---|
| “X” | …PAHLDVLXR430 | …PAHLDVXYR430 |
| A | 4 | 18 |
| V | 2 | 20 |
| I | 3 | 64 |
| L | 3 | 100 (WT) |
| Y | 100 (WT) | 60 |
| F | 4 | 20 |
| W | 6 | 57 |
| R | 3 | 7 |
| D | 5 | 4 |
| C | - | 5 |
| P | - | 7 |
| No PAN | 5 | 5 |
Experiment was performed as described in Table 2
The HbYX motif is required for PAN-20S complex formation
Mutations in the HbYX motif might block gate opening either by preventing the formation of the PAN-20S complex, or by allowing its formation but interfering with the gate-opening mechanism. We used electron microscopy as described previously (Smith et al 2005) to test if the mutations that prevent activation also prevented complex formation. Five different PAN mutants were analyzed by EM. We observed extensive complex formation with those variants that stimulated gate opening, but did not observe any PAN-20S complexes with the PAN mutants that failed to stimulate gate opening (Table S2). Thus, the failure of these PAN mutants to stimulate gate-opening is due to a failure to associate with the 20S.
Residues preceding the HbYX motif are not essential in gating
Although the residues preceding the HbYX motif were not conserved in other proteasomal ATPases, we mutated each of the four residues preceding the HbYX motif to alanines to determine their importance for activation of the 20S. None of these residues was absolutely essential for PAN’s stimulation of peptide hydrolysis, but some alanine replacements did reduce PAN’s activity. Potentially, this C-terminal sequence may adopt a helical conformation as was suggested for the C-termini of PA26 in the PA26-20S complex (Whitby et al., 2000). To test this possibility, we inserted prolines at two positions in the C-termini to prevent helix formation or mutated the proline (P422) to an alanine (Table S3). None of these mutations significantly reduced the stimulatory activity of PAN. Therefore, a C-terminal helix is probably not essential for formation of the PAN-20S complex or activation of gate opening. Also, insertion of four alanines before the seven C-terminal residues had little effect (Table 1), suggesting that these terminal residues are flexible or function relatively independently and fit into a constrained pocket that permits only certain residues.
PAN’s C-terminus moves from an aqueous to a hydrophobic environment upon complex formation
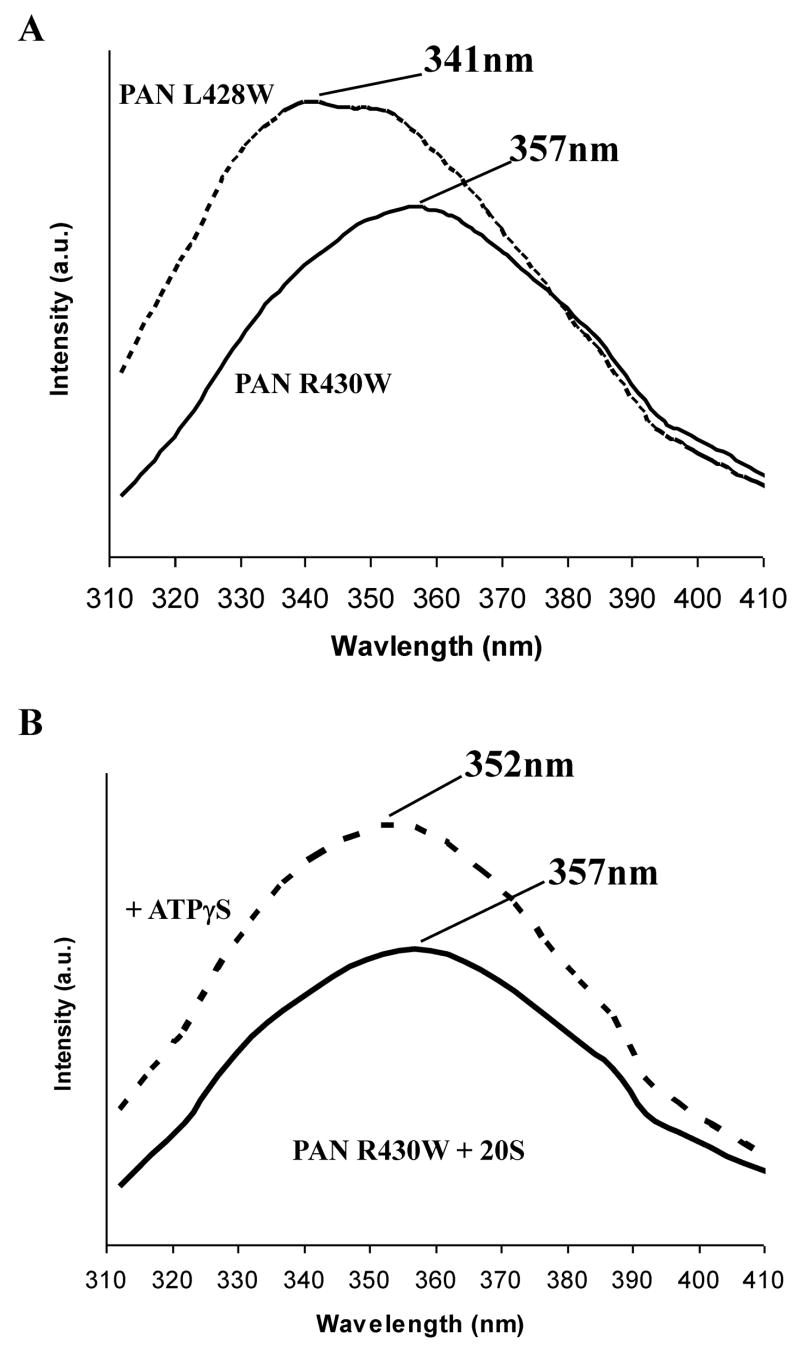
The susceptibility to carboxypeptidase B (Fig. 1C) suggested that PAN’s C-terminal residue is exposed in PAN, but not after association with the 20S. To analyze further the environment surrounding the C-terminal residues, we monitored tryptophan fluorescence. Since wild-type PAN and the Thermoplasma 20S proteasome both lack tryptophans, we analyzed the fluorescence spectra of mutants where a tryptophan was inserted in the C-terminal position (R430) or in place of the conserved hydrophobic residue (L428). Both mutations allowed wild-type activity (Table 1 and 2). The maximum emission wavelength of the C-terminal tryptophan (W430) was 357nm (Fig. 2A), which resembles that of free tryptophan in an aqueous solution. Accordingly, this fluorescence was quenched upon addition of acrylamide (not shown). Therefore, the ultimate C-terminal residue of PAN is indeed exposed to the solvent, where it appears available for docking with the 20S. However, when a tryptophan was placed two residues upstream (W427), it showed a λem max of 341nm, suggesting that this residue is located in a more hydrophobic environment.
Figure 2.
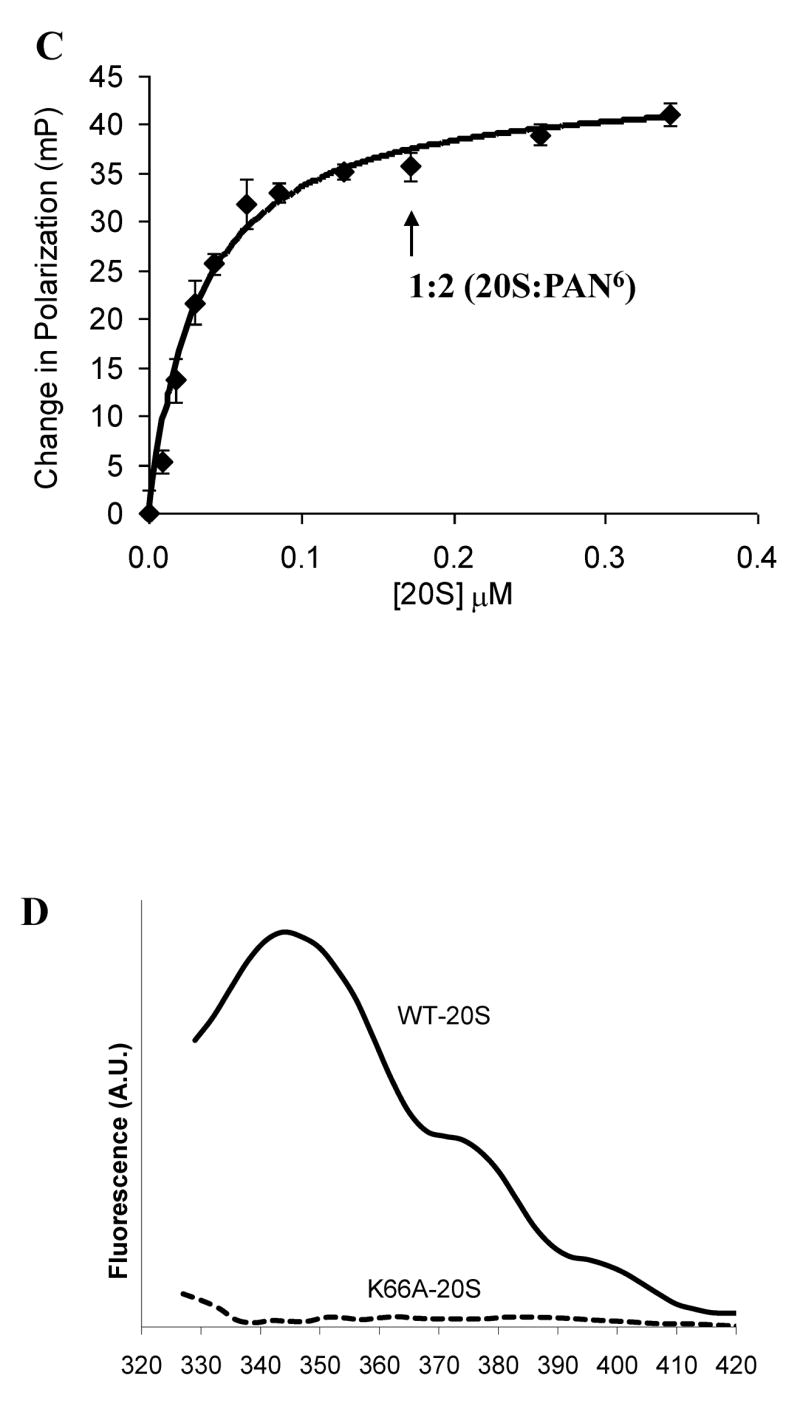
The fluorescent spectrum of PAN R430W and PAN L428W mutants in solution and upon binding the 20S. A) Fluorescent emission spectra of the various PAN mutants. B) Fluorescent emission spectra of PAN R430W in the presence of 20S proteasomes without and with ATPγS. λem-max is indicated. C) Polarization of PAN’s (R430W) tryptophan in the presence of ATPγS upon addition of increasing amount of 20S proteasomes. PAN’s MW was taken as a 585kDa (2-hexameric rings) and 20S as 674kDa. The final concentration of PAN6 was 340nM the 20S concentration is shown. The fit line was calculated from regression analysis of the simple ligand binding equation with one site saturation, the Kd was calculated to be 34 +/−12 nM, and the maximum change in polarization (Bmax) was 45mP. D) A difference plot showing the changes that occur in the fluorescent spectrum of the tryptophan in PAN R430W that is induced by addition of WT-20S or by addition of K66A-20S. The plot was generated by subtracting the spectrum of PAN R430W with the 20S from it’s plot without the 20S, a flat line indicates no change in the spectrum. These experiment were performed at least three times with similar results.
ATP-binding to PAN is essential for complex formation with the 20S (Smith et al., 2005). One attractive model to explain this activation by ATP is that the C-terminal residues assume a more exposed conformation upon binding ATP, which promotes PAN-20S association, but not upon binding ADP, which inhibits complexation. Therefore, we tested whether the tryptophan at the 430 or 428 positions become more or less exposed to solvent upon binding ATP or ADP. However, we found no such change in the spectrum of PAN with either the solvent-exposed R430W or the more hydrophobicly exposed L428W after addition of ATP, ATPγS or ADP.
If the C-termini of PAN, like those of PA26 (Forster et al., 2005), dock into pockets in the 20S’s a-ring upon complex formation, then the terminal tryptophan in the R430W PAN mutant should move from an aqueous to a more hydrophobic environment. After adding ATPγS to a mixture of PAN and proteasomes, the λem max of this terminal tryptophan shifted from 357nm to 352nm, and the fluorescence intensity increased by over 50% (Fig. 2B), as is characteristic of a tryptophan that shifts from an aqueous environment to a more hydrophobic one. By contrast, the addition of ADP did not change the λem max. Also, addition of ATPγS to the 20S (which lacks tryptophans) did not change it’s background tyrosine fluorescence, and there was no change in the spectrum of the PAN L428W mutant upon association, since the λem max of this tryptophan is already blue shifted (Fig 2A). At high concentrations, ADP competes with ATP and promotes PAN-20S dissociation (Smith et al., 2005). Accordingly, if excess ADP was added after formation of the PAN-20S complex with ATPγS, then ADP shifted the λem max to the red, and the fluorescence intensity decreased (not shown). Therefore, upon ATP-dependent association of PAN with the 20S, the C-termini move from an aqueous environment to a more hydrophobic one, presumably the pockets in the α-ring (see below).
PAN’s C-termini become polarized upon association with the 20S
Formation of the PAN-20S complex has been difficult to demonstrate because of its transient, and relatively weak association (Smith et al., 2005). To quantitate the association of PAN with the 20S in solution, we analyzed the polarization of the C-terminal tryptophan in PAN R430W. Fluorescence polarization can be used to measure the rate of rotation of a fluorescent molecule, which can decrease upon association with another protein. When ATPγS (but not ADP) was added to a solution of PAN and the 20S, a 40 mP change in polarization was detected. Also, without the 20S present, W430 did not become more polarized upon addition of ATP, ATPγS or ADP. Thus, the terminal tryptophan becomes polarized upon association with the 20S. To determine what molar ratio of 20S to PAN gave maximal polarization, we added increasing amounts of 20S to a solution of PAN R430W and ATPγS. This assay is more quantitative than biochemical or EM approaches used previously to show complex formation, and because high concentrations of PAN and 20S could be used, stochiometric binding and saturation were demonstrated. Polarization approached maximal with two hexameric PAN complexes per 20S proteasome (Fig. 2C), i.e. when most of the 20S was doubly capped with PAN, and most of the PAN was associated with a proteasome.
PAN binding to the 20S requires lysine-66 in its α-ring
Lysine 66 is located in the intersubunit pockets in the 20S’s α-ring, and when PA26 complexes with the Thermoplasma 20S, the C-terminal carboxyl group of PA26 forms a hydrogen bond with this lysine (Forster et al., 2005). Accordingly, we found that PAN was unable to stimulate gate opening in K66A-20S mutant proteasomes, as assayed by measuring LFP hydrolysis (data not shown). Forster et. al. (2005) had shown that this lysine is also required for the stimulation of casein degradation by PAN, which could be due to the requirement of Lys66 for gate-opening or for PAN-20S association. We therefore tested if K66 is also essential for the association of PAN and the 20S. In the presence of ATPγS, the K66A-20S, unlike wild-type proteasomes, did not alter the fluorescence spectrum of the terminal tryptophan in PAN-R430W (Fig 2D) and did not cause its polarization (data not shown). These results indicate that PAN cannot stimulate gate opening in K66A-20S because it cannot associate with this particle and strongly suggest that PAN’s C-termini dock into the same intersubunit pockets as the C-termini of PA26.
Short peptides corresponding to PAN’s C-terminus induce 20S gate opening
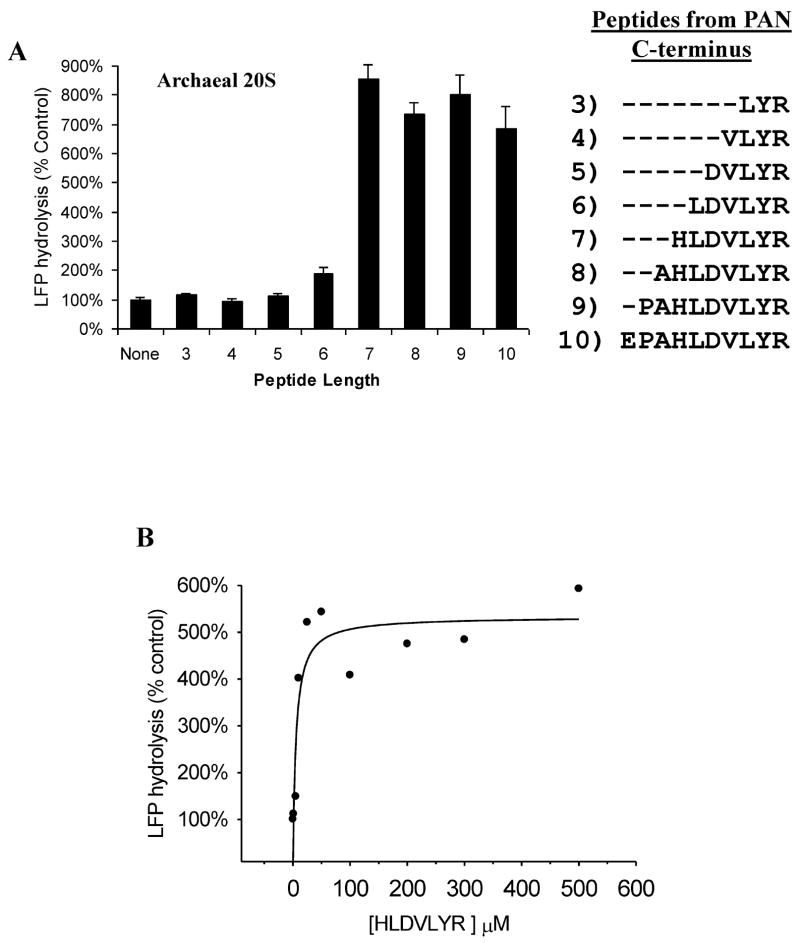
Two possible models can explain the above results: 1) that insertion of PAN’s C-termini into the hydrophobic binding sites in the α-ring function by themselves like a “key-in-a-lock” to induce gate opening. 2) Alternatively, this interaction is necessary for PAN-20S complexation, but some other domain(s) in PAN (e.g. like the activation loop in PA28/26) are also necessary to induce the open-gate conformation. To test whether docking of PAN’s C-termini is sufficient to cause gate opening, we synthesized different length peptides that corresponded to PAN’s C-terminal sequence (Fig. 3A), incubated them with proteasomes, and monitored gate opening by LFP hydrolysis. Surprisingly, peptides 7 to 10 residues long by themselves stimulated peptide degradation by 20S proteasomes by up to 9-fold. This large stimulation, which resembled the stimulation seen with PAN and ATP, was seen at the rather high peptide concentrations of 250 μM (Fig. 3A). However, with the 7 residue peptide, near-maximal stimulation of peptide hydrolysis was evident at 10μM (See supplementary, Fig. S2). These findings indicate rather high affinity binding of the 7-residue peptide, especially since in PAN, these C-termini are likely to function in a multimeric fashion and with a specific steric relationship. However, accurate determination of Ka’s are unreliable by this approach, because these C-terminal peptides (especially above 50μM), not only cause gate-opening, but also enter the 20S particle and compete at the active sites with LFP (whose hydrolysis is used to assay gate opening).
Figure 3.
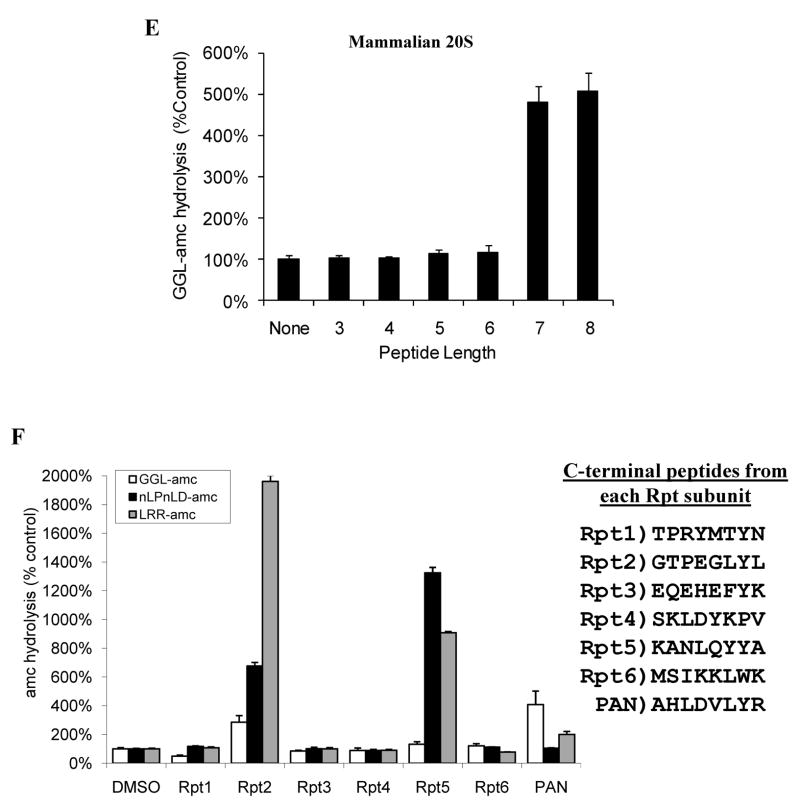
Peptides derived from PAN’s extreme C-terminal sequence induce gate opening and inhibit PAN-20S complex assembly. A) Ability of peptides of different lengths to stimulate gate opening. Peptides (250μM) were incubated with 0.2 μg 20S proteasomes and LFP. In each case, a correction was made because the added peptide, in addition to causing gate opening, competed with LFP at the active sites. To determine the actual percentage stimulation of LFP hydrolysis due to gate-opening, the ability of the various peptides to inhibit LFP hydrolysis by the gateless (Δα2–12) 20S proteasome was also measured, and the values used to normalize data on LFP hydrolysis by WT 20S proteasomes. Without normalization, 7-8 residue peptides still stimulated 20S proteasomes 3–4 fold (see also B). Values in A, C, D and E are the means of three independent experiments +/− SD’s. B) The 7 residue peptide from PAN’s C-terminus (HLDVLYR) induces gate opening in the Thermoplasma 20S at much lower concentrations than the 10 residue peptide from PAN’s C-terminus (EPAHLDVLYR). This experiment was performed as in A) using the concentrations of the peptide shown, except that no adjustment for competition with the fluorogenic substrate was calculated, since such corrections become unreliable at the high peptide concentrations (>500 μM). The peptide HLDVLYR inhibits cleavage of LFP by Δα(2–12)20S at ≥50μM. C) Replacement of the Hb (hydrophobic) and Y residues in the 8-residue peptides prevented gate-opening. Proteasome activity was measured as in A) in the presence of the indicated peptides. Also shown is the inability of the WT peptide (250μM) to induce gate opening in the 20S K66A, and the inability of the peptide corresponding to PA26’s C-termini (500 SM)* to induce gate opening in wild-type 20S. D) Peptides that activate gate-opening prevent association of the 20S and PAN, as does the peptide corresponding to the C-terminus of PA26. Each peptide was pre-incubated with 20S and PAN for five minutes followed by addition of ATPγS. Polarization was measured before and after nucleotide addition. No peptide (control) was taken as 100% polarization and was similar to that shown in Fig. 2. The values are means +/− SD’s from at least three experiments. Peptide concentrations were used as in C). E) A similar experiment as in A (using peptides corresponding to PAN’s c-terminus) was carried out using purified rabbit muscle 20S proteasomes and GGL-amc as the fluorogenic substrate (Smith et al., 2005). F) A similar experiment as in C was carried out using purified rabbit muscle 20S but with peptides that corresponded to the C-termini of the mammalian 19S ATPases: Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6. The cleavage rates of fluorogenic substrates specific to its three peptidase sites were monitored (see text). The peptide corresponding to Rpt1 stimulated gate opening weakly under some conditions (e.g. no glycerol but with 60mM KCl). The sequences for each of the C-terminal peptides are shown. All values are the Means +/− SD of three independent experiments.
The 7-residue peptide, HLDVLYR, stimulated gate opening much better than the 6-residue peptide, LDVLYR, and smaller peptides were ineffective. The small stimulation with the 6-residue peptide (up to 2-fold) reflects a requirement for peptide length rather than for an N-terminal histidine, since when this histidine was changed to an alanine in the peptide, it showed a similar ability to stimulate gate opening. Accordingly the corresponding mutation in full length PAN, A424H, stimulated gate opening like wild-type PAN. Surprisingly, the efficacy of the C-terminal peptides decreased at greater length, and the 10-residue peptide (EPAHLDVLYR) showed activity only above 200μM (data not shown). These findings indicate a steep dependence on peptide length. Presumably, a length of 7-residues is necessary for the peptide to occupy and assume a similar conformation as the C-termini of PAN in the intersubunit pockets.
Peptide-induced gate opening requires the HbYX motif and lysine-66 in the α-ring
Gate opening in the 20S can be activated non-specifically by detergents, high temperature, and some hydrophobic peptides (Coux et al., 1996). To determine if the 7-residue peptides corresponding to PAN’s C-terminus cause gate-opening by the same mechanism as PAN, we tested whether the peptide must also contain the HbYX motif. When the Hb or the Y residue was replaced by an alanine, the peptide was not able to stimulate gate opening, as was found with PAN (Table 2). Also, replacement of the C-terminal arginine with an alanine did not reduce its activity, as was found with the corresponding mutation in PAN (Table 1). Furthermore, the 7-residue peptide from PAN’s C-terminus could not stimulate gate opening in proteasomes expressing the K66A mutation in the α-subunits (Fig. 3B). Thus, gate opening by the C-terminal peptides and PAN shows the same requirements for an HbYX motif and a lysine 66 in the intersubunit pockets, indicating that they function by very similar mechanism.
Peptides from PAN’s C-terminus inhibit PAN-20S association
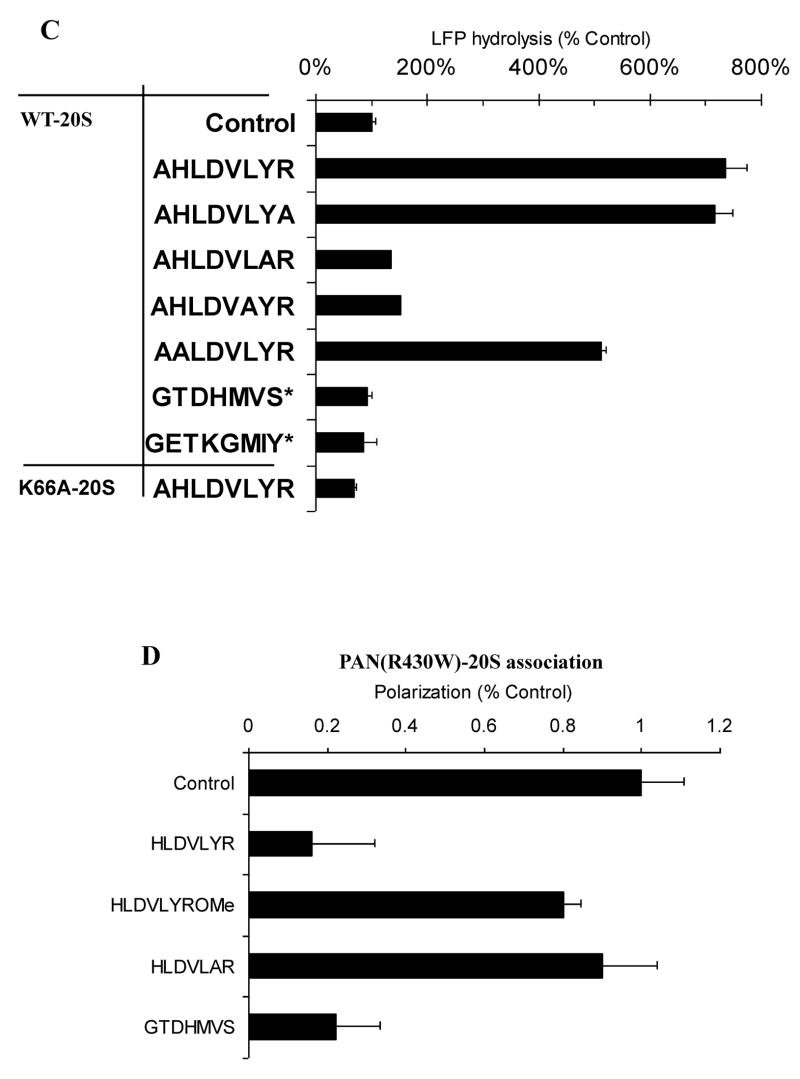
If these peptides induce gate opening by docking into the same sites in the α-ring as PAN’s C-termini, then these peptides should act as competitive inhibitors of the PAN-20S association. We therefore pre-incubated these peptides (250SM) with 20S proteasomes and R430W PAN (350nM) for five minutes prior to the addition of ATPγS, and then monitored complex formation by measuring polarization of PAN’s terminal tryptophan. The wild-type peptide inhibited complex formation nearly completely throughout the course of the experiment, while the inactive peptide containing an alanine in place of the conserved tyrosine did not inhibit (Fig. 3C).
Since an additional alanine on PAN’s C-terminus prevents complex formation (Table S2), it seemed likely that a free carboxyl group in this position is important for binding to the 20S. To test this possibility, we blocked the C-terminal carboxyl group on HLDVLYR by esterification with methanolic acid. After carboxy-esterification, the peptide completely lost its ability to inhibit polarization (Fig. 3C). Thus a carboxyl group is essential for docking in the α-ring. Together, these observations establish that gate opening induced by peptides from PAN’s C-terminus is a specific response that requires a length of 7 residues, the conserved HbYX motif, a free carboxyl group and lysine 66 in the 20S’s α-ring. In related studies, we have used cryo-electron microscopy to confirm that the gate-opening peptides bind to the intersubunit pockets in the 20S’s α-ring adjacent to lys66 (Rabl et al, Ms in Preparation).
C-termini of PA26 bind but do not cause gate opening
Because the C-termini of PAN and PA26 both require lysine 66 in the α-subunits to associate with the 20S and activate gate opening, they are likely to bind to or act through the same sites (Forster et al., 2005), even though PA28/26 lack the HbYX motif. Therefore, we studied under these same conditions the effects on gating of the seven residues peptide corresponding to the C-terminus of PA26, GTPHMVS, or eight residues peptides from the C-terminus of PA28α (which by itself forms a heptameric complex that induces gate opening in eukaryotic 20S proteasomes (Cascio et al., 2002)). However, in contrast to the 7- or 8-residue peptides from PAN’s C-terminus, these peptides could not by themselves induce gate-opening at 250 μM (Fig. 3A) or even at 1mM (not shown). The failure of this PA26 peptide to stimulate gate opening is consistent with the prior findings that the C-terminal residues of PA26 are required for its association with the 20S, but that this binding does not induce gate opening, which requires the distant activation domain of PA26 (Zhang et al., 1998).
To determine whether the heptapeptide from PA26 actually binds to the 20S, we measured fluorescence polarization of PAN R430W to test whether this C-terminal peptide could competitively inhibit PAN’s association with the 20S. After preincubation with the 20S, this peptide, like those from PAN’s C-terminus, was found to inhibit the polarization that was associated with PAN-20S complex formation (Fig. 3D). However, this inhibition of polarization, unlike that by the C-terminal peptides from PAN, was temporary and lasted for only several minutes, presumably because the PA26-derived peptide had much lower affinity for the 20S than PAN and the PAN-derived C-terminal peptide.
C-terminal peptides from PAN and certain 19S ATPases induce gate opening in the mammalian 20S
Since the HbYX motif is also found in the C-termini of three of the ATPases in the 26S proteasome, it seemed likely that these C-termini regulate gating in the eukaryotic 20S in a similar fashion as PAN. We therefore tested if similar mechanisms function in the 26S particle, and if short peptides from PAN’s C-terminus could induce gate opening in proteasomes from rabbit muscle. In these particles, gate opening was monitored with the tripeptide substrate GGL-amc, which is excluded by the closed gate conformation. The 7 and 8 residue peptides from PAN were indeed able to cause a 5–6 fold increase in peptide entry, but smaller ones had no effect. Thus, the mammalian 20S showed the same length requirement for gate opening as did the archaeal 20S (Fig. 3A and E).
To determine whether the ATPases in the 26S proteasome function similarly to PAN, we synthesized eight-residue peptides corresponding to the C-terminus of each of the mammalian 19S ATPases, Rpt1–6. Because such C-terminal peptides may also enter the 20S particle and compete with the fluorogenic substrates for binding to the active sites, we monitored the induction of gate opening with substrates specific for its different catalytic sites (i.e. chymotrypsin-like site: GGL-amc, caspase-like site: nLPnLD-amc, and trypsin-like site: LRR-amc) to reduce the chances that substrate competition would mask the effects on gate opening. The C-terminal peptides from Rpt2 and Rpt5, which contains the HbYX motif, strongly stimulated gate opening in the rabbit 20S proteasome, and did so to a greater extent than the C-terminal peptide from PAN (Fig. 3F). Prior genetic studies had also implicated Rpt2 in regulation of gate-opening (Kohler et al, 2002). As expected, Rpt 4 and Rpt6 which do not contain the HbYX motif, could not induce gate opening. Rpt1 was found to induce gate opening weakly but only under some conditions, possibly because the residue preceding its penultimate tyrosine is a threonine (not a hydrophobic residue) and thus does not conform to the HbYX motif. Surprisingly, Rpt3, whose last three residues (…FYK) does contain the HbYX motif was never found to induce gate opening in mammalian 20S particles. Related observations indicated that lysines in the C-terminal (X) position do not support gate opening in mammalian 20S in contrast to the HbYX motif in PAN which functions with basic (but not acidic) residues in this position (DS and AG, data not shown). Thus, gate opening by the HbYX motif appears to function in 26S proteasomes, although gating seems to be regulated directly by only C-termini of Rpt2 and Rpt5, although other C-termini may contribute indirectly (e.g. by stabilizing the 26S complex) (see below).
The mammalian 19S ATPases’ C-termini appear to be essential for gate opening
To determine if the C-termini of the 19S ATPases, Rpt1–6, are also required for gate opening in mammalian 26S, we studied the effects of carboxypeptidase treatment of purified mammalian 19S/PA700 complex (kindly provided by George DeMartino) on its ability to induce gate opening in purified rabbit 20S proteasomes, in an experiment similar to that done with PAN (Fig. 1C). The C-terminal residues of the ATPases Rpt3 and Rpt6 are lysines which should be removed by carboxypeptidase B, while those on Rpt2 (Leu), Rpt4 (Val), and possibly Rpt5 (Ala) should be removed by carboxypeptidase A. Treatment of the 19S complex with either carboxypeptidase A or B prior to addition of the 20S and ATPγS significantly reduced the ability of the 19S to stimulate peptide hydrolysis in the presence of ATPγS (Fig. 4A). Thus, both classes of 19S C-termini seem important for the stimulation of gate opening. (In these experiments, carboxypeptidases B seemed more effective than A, but these effects cannot be compared quantitatively since the extent of removal of the different C-terminal residues by the two enzyme preparations may well differ.)
Figure 4.
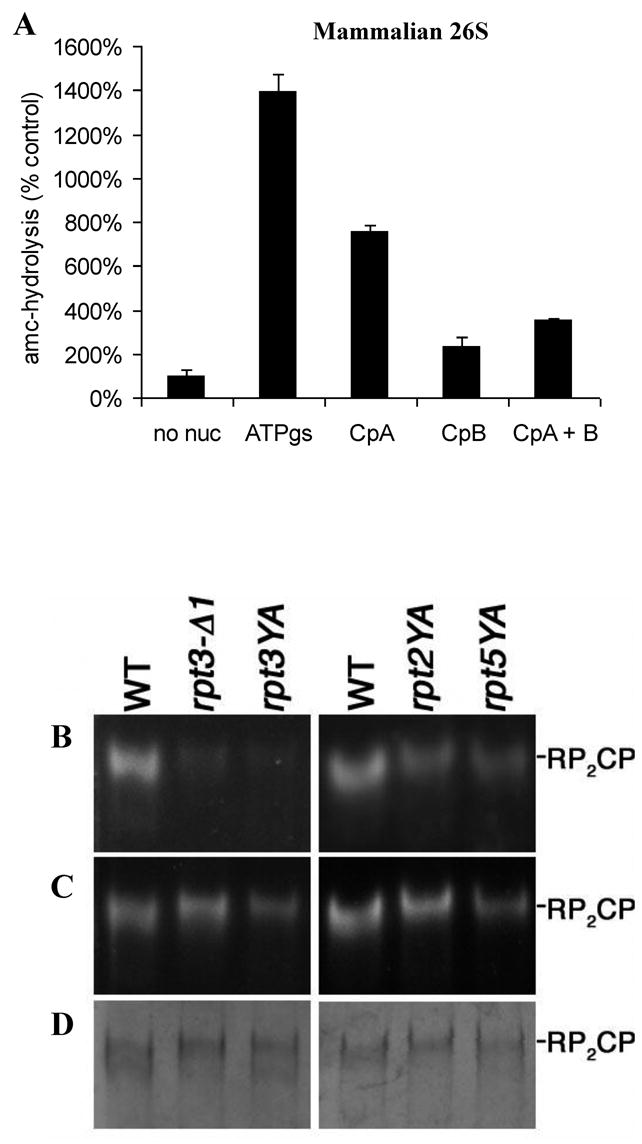
Carboxypeptidase induced truncation and mutations within the HbYX motif result in 26S instability and gating defects. A) Carboxypeptidase treatment of free mammalian 19S/PA700 inhibits its ability to stimulate peptide hydrolysis by the mammalian 20S in the presence of ATPγS. The 19S particles (1μg/reaction) were preincubated with or without 0.1 U of carboxypeptidase A or B for 10 min. prior to addition of the 20S (0.1μg/reaction) and ATPγS (0.1mM). The assembly reaction mix was diluted 5 fold in the reaction buffer with Ac-nLPnLD-amc and the rate of hydrolysis was monitored. B) Purified proteasomes (3μg) from wild-type (WT) and rpt mutant strains were resolved on 3.5% nondenaturing gels, and proteasome activity was visualized by staining with the fluorogenic substrate suc-LLVY-AMC. The YA mutants are single-residue substitutions of the penultimate Tyr of the indicated Rpt protein. rpt3-Δ1 is a deletion of the C-terminal lysine of Rpt3. C) After the initial peptidase assay in (A), the assay was repeated in the presence of 0.02% SDS to cause gate opening. D) The native gels were stained with Coomassie Brilliant Blue to assess the amount of protein in each sample.
By contrast, treatment of the isolated 26S complex with carboxypeptidase A or B did not reduce the stimulation of peptide hydrolysis by ATPγS (not shown), presumably because the critical C-terminal residue in the 19S ATPases are also buried upon complex formation, as was found with the PAN-20S complex. These latter observations also indicate that the carboxypeptidase treatments did not destroy the integrity of the 19S complex or act indirectly by affecting subunits outside the 19S–20S interacting surfaces.
The C-terminal YX residues in the yeast 26S ATPases are required for gate opening
These findings suggest that the C-termini of multiple ATPase subunits are important in 26S formation and gating. To further test whether the terminal YX residues are necessary for 26S stability or gate-opening, we examined the effects of mutagenesis of the YX residues in the C-termini of the four ATPase subunits in S. cerevisiae proteasomes. We substituted an alanine for the conserved penultimate tyrosine in the four ATPase subunits that contain this residue, Rpt1, Rpt2, Rpt3, and Rpt5, and purified the 26S proteasomes from these mutant strains using affinity tags (Leggett et al., 2002). It is noteworthy that each of all these YA replacements in the four ATPase C-termini caused highly complex and distinct complex phenotypes related to protein degradation, but all allowed sufficient proteasome function for growth of the yeast (S.P. and D.F., unpublished data). Analysis of these phenotypic defects is beyond the scope of this study and will be published elsewhere. The activity of each mutant proteasome was assessed in nondenaturing gels. After electrophoresis, the gels were incubated in the presence of the fluorogenic substrate, suc-LLVY-amc, to assess the extent of complexation and gate opening (Fig. 4B). Replacement of the conserved tyrosine in Rpt1 by an alanine appeared to cause a major defect in the stability of the 26S complex, since we were not able to isolate these particles by standard affinity purification methods. By contrast, the rpt2YA, rpt3YA, and rpt5YA mutant proteasomes were successfully purified by these approaches, but they all exhibited reduced suc-LLVY-amc hydrolysis. These defects in peptide hydrolysis reflect primarily decreased substrate entry and not simply reduced 26S proteasome levels, because equivalent proteasome loading was evident when the gels were stained, and the 26S proteasomes did not disassemble during electrophoresis or during incubation with substrate (see supplement and Fig. S2). In addition, to test the capacity for gate opening, this assay was repeated in the presence of 0.02% SDS, which artificially induces gate opening (Fig 4C). The rpt3YA and rpt2YA mutant proteasomes showed a significant increase in peptide hydrolysis with SDS, but the rpt5YA mutant showed only a limited response. Thus, the conserved tyrosines in each of the four ATPases appear important for regulating gate opening in the 20S particle, but their specific roles appear to differ.
A similar approach was then used to delete the C-terminal residues on the six 19S ATPases. Each of these mutants also had phenotypic defects, related to proteolysis, which will be analyzed elsewhere. The C-terminal deletion in Rpt3 resulted in a clear gating defect similar to that seen with the Tyr to Ala substitutions in Rpt2, Rpt3 and Rpt5 (Fig 4C). When the C-termini of the other ATPases Rpt4, Rpt5, and Rpt6, were truncated by a single C-terminal residue, there appeared to be stability problems in the 26S complex that reduced recovery (S.P. and D.F. unpublished data). Thus, the terminal YX residues are necessary for proper gating in the yeast 26S proteasome, and some appear necessary for its stability. While the terminal HbYX motifs in the 19S ATPases appear to play important functions in gating and complex formation that are consistent with the findings on PAN, the specific roles of the individual ATPases C-termini clearly differ from one another.
Discussion
The ATPases’ mechanism for activation of the 20S
The present studies have uncovered an essential new ATP-dependent step in proteasome function and have clarified how the regulatory ATPases facilitate substrate entry into the 20S proteasome. When ATP binds to the proteasomal ATPases, the conserved HbYX motif in their C-termini dock into pockets in the 20S α-ring and function like a “key-in-a-lock” to open the gate that limits substrate entry (Fig. 5A). Because 7-residue peptides corresponding to the C-termini of either PAN, Rpt2 or Rpt5 by themselves bind and trigger gate opening in both archaeal and mammalian proteasomes (Fig. 5B), this novel reaction does not require other parts of the ATPase molecule and appears to represent a general mechanism regulating proteasome function. The HbYX motif is present in the C-termini of PAN found in most species of archaea, and in three of the 26S ATPases (while four of the ATPases contain a penultimate tyrosine) in all eukaryotic species where sequences are available. It is noteworthy that similar C-terminal sequences are also present in the presumed proteasome-regulatory ATPases in rhodococcus and mycobacteria (Darwin et al., 2003). In fact in-vivo studies have demonstrated a two-residue truncation of this motif in the proteasomal ATPase (MPA) in Mycobacterium tuberculosis results in a conditional lethal phenotype (Darwin et al., 2003). In PAN from several archaeal species, there is a HbFX in place of the conserved HbYX (Table S1). Presumably, in these species, the structure of the intersubunit pockets in the 20S α-rings allows selective binding and gate-opening by this alternative HbFX motif.
Figure 5.
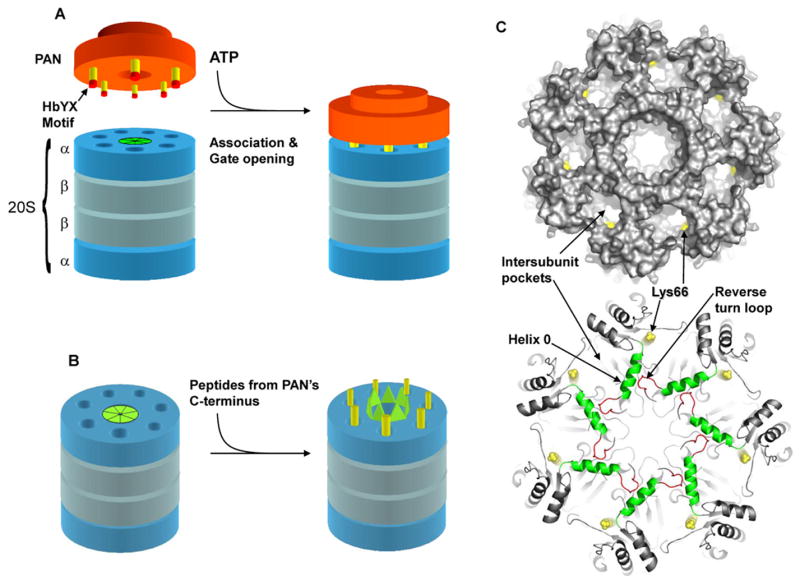
A) Model depicting the association of PAN with the α-ring of the 20S proteasome, in which the C-termini (yellow) of PAN (orange) dock into the intersubunit pockets in the top of the 20S. B) Schematic model for gate opening in the 20S upon binding of peptides derived form PAN’s C-terminus to the intersubunit pockets in the 20S. C) Top: top view of the 20S α-ring with a surface rendering to demonstrate the intersubunit pockets and location of lys66 (yellow). Bottom: Ribbon representation of the 20S α-ring. Peptide binding to lys66 (yellow) adjacent to Helix 0 (green) may cause a conformational change in Helix 0 that propagates to the reverse turn loop (red) to induce gate opening. The N-terminal gating residues are not resolved in this crystal structure (1PMA). Images in C) were rendered with Pymol.
No HbYX or related C-terminal motif is present in the bacterial ATPase complexes (ClpA, ClpX, ClpC and HslU) that activate the ClpP or HslV peptidase complexes, and their modes of activation of proteolysis differ from that demonstrated here. Unlike PAN, the regulatory Clp ATPases associate with ClpP via an internal loop, instead of their C-terminal sequences (Guo et al., 2002). A more similar mechanism for ATP-dependent activation occurs with HslU, which is more homologous to PAN and the 19S ATPases. Upon ATP-binding to HslU, its six C-termini dock into the six crevices in the HslV peptidase complex and induce conformational changes that enhance its proteolytic activity (Seong et al., 2002). However, removal of up to five residues from the C-terminus of HslU does not block its ability to associate with and stimulate HslV (Seong et al., 2002). Thus HslU does not require a specific C-terminal motif for its function, which is in contrast to our findings with the proteasomal ATPases. Furthermore, both ClpP and HslV are 2-ring complexes that lack an outer ring or a gated pore formed by the subunits’ N-termini, and thus their ATP-dependent mechanisms of activation, which occurs through alteration in the proteolytic sites, are fundamentally different from gate opening in the a-ring of the 20S proteasome.
Exactly how binding of ATP or ATPγS to PAN allows its C-termini to dock into the intersubunit pockets is unclear. The simplest mechanism would be that the critical HbYX motif is buried in the ADP-bound state, but becomes exposed upon binding of ATP or ATPγS. ATP-induced exposure of C-terminal residues for docking has been demonstrated by Chung and coworkers (Wang et al., 2001) for HslUV from E. coli, which is a 6-fold symmetric complex unlike the ATPase-proteasome complex. However, our measurements of tryptophan fluorescence and susceptibility to carboxypeptidase B indicate that upon binding of ATP or ADP, PAN’s C-termini do not shift between buried and exposed states but instead remain in an exposed, aqueous environment until they associate with the intersubunit pockets in the 20S particle’s α-ring. Although PAN’s C-termini do not appear to transition between buried and exposed states to promote complex formation, ATP-driven changes in their conformation within the aqueous environment may promote association by altering their spacing to allow a better fit with the docking sites on the 20S. Alternatively, PAN may contain blocking domains that become buried upon ATP binding, allowing the C-terminal regions to dock in the α-ring.
Because of its key role in allowing proteolysis, the binding of the HbYX motif to these intersubunit pockets for gate opening represents an attractive drug target. Proteasome inhibitors, which block some of the peptidase sites within the 20S’s central chamber, have found wide application as research tools and in the treatment of certain cancers (Adams, 2004; Kisselev and Goldberg, 2001). Inhibiting the initial binding of the C-termini to the α-ring should inhibit 26S assembly, substrate entry into the 20S proteasome, and protein degradation.
The gate-opening mechanisms of the ATPases and PA28/26 are distinct
Although gate opening by the ATPases and PA28/26 have been proposed to occur by similar mechanisms (Forster et al., 2005; Forster et al., 2003; Kohler et al., 2001), this study demonstrates fundamental differences. Unlike the peptides from PAN’s C-terminus, the C-terminal 7-residue peptide from PA26 or the 8-residue peptide from PA28α could not induce gate opening (Fig. 3B and C) even at 1mM concentrations. Their inability to stimulate was expected, since gate-opening by PA28/26 requires an activation loop, distant from its C-termini (Zhang et al., 1998). When this activation domain is mutated, PA28 can still stably associate with the 20S, and its C-termini can dock into the intersubunit pockets in the α-ring, but it cannot activate gate opening. Thus, for PA28 (unlike PAN) binding to the α-ring and gate opening can be dissociated. Since PAN’s C-terminal sequence alone induces gate opening, it clearly does not require such an activation domain and can work at relatively low concentrations (≤10μM). The activation domain in PA26 induces gate opening by pressing on and displacing the reverse turn loop located between the N-terminal gating residues and the contiguous Helix 0 (see Fig. 5C and (Whitby et al., 2000)). This gating mechanism requires seven equilateral C-terminal residues and a distant activation domain in a specific rigid three-dimensional framework. However, a 7-residue peptide does not have the structural capacity to function by such a mechanism, and it is too small to interact with additional domains outside the intersubunit pockets. In addition, PAN’s ability to induce gate opening was preserved even with a four alanine extension preceding the last 7-residues of PAN, and such an insertion should prevent a direct interaction between any upstream “activation loops” and the surface of the α-ring. Our related cryo-EM studies (Rabl et. al. In Preparation) demonstrate that upon binding, these HbYX containing peptides (unlike C-terminal peptides from PA26) induce a novel conformational change in each α-subunit, which results in a displacement of the Pro17 reverse turn loop in a manner that stabilizes the open gate conformation.
Despite having different consequences, the binding of PAN’s and PA26’s C-termini appear to occur in the same pockets, since the peptides from both can inhibit the binding of PAN to the 20S complex. Also, gate opening by both requires lysine 66 in the α-subunits (Fig 5C top). PA26’s C-terminal carboxyl group forms a hydrogen bond with lysine 66, and we have confirmed the prior suggestion (Forster et al., 2005) that this lysine is important for the PAN-20S interaction. Accordingly, gate-opening by the octapeptide from PAN’s C-terminus required a free C-terminal carboxyl group. Therefore, though the C-termini of PAN and PA28/26 share a common binding site on the 20S the consequences of binding depends on the presence of the HbYX motif.
Distinct mechanisms for PA28/26 and the proteasomal ATPases should not be surprising since they lack sequence homology throughout, especially in their critical C-termini, and differ in their quaternary structures and their in vivo functions. Furthermore, the imposing of PA28/26’s 7-fold symmetry onto the 20S’ α-ring is of fundamental importance to its mechanism of gate opening (Forster et al., 2003; Whitby et al., 2000). By contrast, the proteasomal ATPases have a 6-fold symmetry, which implies that only a fraction of the 20S 7-intersubunit pockets can be occupied by the C-termini of the ATPases, and because of geometric constraints, it is likely that only a few of the ATPases C-termini can possibly bind at any one time. Thus, PAN and the 19S complex cannot impose a 7-fold symmetry on the α-subunits. A longstanding fundamental puzzle is how a 6-subunit ATPase ring complex can associate with the 20S and stabilize its’ 7-fold symmetric open-gate conformation. The only solution is that gate opening by the ATPases requires docking of only a subset of C-termini to the α-intersubunit pockets, probably only two to four termini, depending on the extent of C-terminal flexibility.
Such asymmetrical contact between the ATPases and the 20S α-ring is supported by electron microscopy of both the 26S (Walz et al., 1998) and the PAN-20S complex (Smith et al., 2005), which demonstrates that often only single small sections of the ATPase ring closely contact the 20S α-ring, unlike PA28/26, which makes flush contact with the 20S. This apparent “wobbling” of the ATPase on the 20S may reflect the docking of only a subset of the C-termini to the 20S α-ring at any time. In addition, several homologous hexameric ATPases (e.g. p97, HslU, ClpX) have been shown to bind ATP to only 2–4 subunits at any time (Bochtler et al., 2000; DeLaBarre and Brunger, 2005; Hersch et al., 2005; Singleton et al., 2000), and PAN behaves similarly (Smith, Reis, and Goldberg, unpublished observations). Since ATP binding is required for PAN-20S association, it seems likely that only the C-termini from the ATP-bound subunits are docked at any instant, and that the interacting subunits alternate as the different subunits are in their ATP or ADP-bound forms (Smith et al., 2005). Like other members of the AAA family of ATPases, PAN and the 19S ATPases must undergo extensive ATP/ADP dependent conformational changes, and are thus highly dynamic structures (DeLaBarre and Brunger, 2005; Rouiller et al., 2002; Wang et al., 2001). By contrast, PA28/26 do not exhibit such dynamics or flexibility, which may help explain their different gate-opening mechanisms. In fact, the function of the “activation loop” requires that PA28/26 be a rigid structure with a symmetric, multivalent, and flush interaction involving a 7:7 matched interface with the α-ring. By contrast, the highly dynamic structure of the ATPases, whose conformational changes drive protein unfolding and translocation, probably requires the C-terminal “key-in-a-lock” mechanism described here and a flexibly tethered 6:7 mismatched interface, which is probably the only type of mechanism by which an ATPase complex can undergo major conformational changes and simultaneously stabilize the open-gate conformation of the α-subunits.
The HbYX motif functions in complex formation and gating in the 26S proteasome
Asymmetric binding of the ATPases’ C-termini to only some of the intersubunit pockets clearly must be functioning in the 26S complex, because the six ATPases in the base of the 19S complex are not identical molecules, and two lack an a penultimate tyrosine, and are thus likely to interact quite differently with the 20S. In addition, the conformational arrangement of the gate in the eukaryotic 20S is highly asymmetric. Only three of the α-subunits N-termini (i.e. α2, α3 and α4) stabilize the closed gate conformation with α3 being the central player (Groll et al., 2000), and removal of only one N-terminus from the 20S’s α-subunits (α3) is sufficient to open the gate (Kohler et al., 2001). In addition, mutation in the ATP-binding domain of a single ATPase subunit, Rpt2, blocks gate opening in the yeast 26S (Kohler et al., 2001), and we show here that the C-terminal peptides only from Rpt2 or Rpt5 are able to stimulate gate opening in the mammalian 20S proteasome. Although we do not know how many pockets these peptides bind to, the docking of only one C-terminal HbYX motif into one specific intersubunit pocket may be sufficient to alter the conformation of the α3-subunit’s N-terminus and to induce gate opening. This finding that docking by one (or two) HbYX motifs is sufficient for gate-opening is in agreement with the steric restriction that only a limited number of the six C-termini can interact with the seven intersubunit pockets. Moreover since single point mutations in the C-terminal YX of Rpt 2, Rpt3, or Rpt5, can inhibit gate opening, while still permitting 26S complex formation, the ATPases also must not contain distinct “activation domains”, like that in PA28/26.
The processes of 19S–20S association and gating are clearly much more complex than in the PAN-20S complex, due to the presence of six different ATPases with distinct C-termini and seven distinct intersubunit pockets. Nevertheless, the “key-in-a lock” mechanism seems to be conserved from archaea to mammalian proteasomes as shown by carboxypeptidase treatments and mutagenesis, and the C-terminal YX residues appear to be critical for 26S formation and gating. The removal of these C-terminal residues from single subunits reduces 26S stability or gating in yeast, and loss of these C-termini from multiple subunits in mammalian proteasomes by carboxypeptidase A or B blocks 20S gate opening by the 19S complex.
It is also evident from these results that the individual 19S ATPase’s C-termini have distinct functions and may be important in complexation, in gating, in both or in neither. For example, the loss of the penultimate tyrosine interferes markedly with 26S stability in the case of Rpt1, while this mutation in Rpt2, 3, and 5 generates fairly stable complexes but with significant gating defects. Furthermore, only the C-terminal octapeptides from Rpt2 and Rpt5 appear capable of triggering gate opening by themselves, and mutagenesis of the ATP binding sites on the different ATPases had indicated a special role of Rpt2 in the gating process (Kohler et al., 2001). While a mutation in PAN’s C-terminus alters all six of it’s C-termini similarly, it is quite surprising and impressive that a single substitution in only one of the six different ATPases in the 19S still causes gating defects and can destabilize the complex. Also, it is noteworthy that point mutations in Rpt3’s C-terminus prevented gate opening in yeast proteasomes even though a C-terminal peptide from Rpt3 cold not induce gate-opening by itself. These observations suggest that while some C-termini play specific roles (e.g. rpt2 and 5 in gating) these actions may require the function of other neighbouring C-termini (e.g. rpt3’s association with an intersubunit pocket). It is thus likely that the ATP-dependent docking of the C-termini of some ATPases to specific pockets in the 20S may be important for 26S complex formation and gating by orienting the ATPases on the α-ring so as to facilitate gate-opening by Rpt2 and Rpt5.
The extensive conservation of these ATPase subunits and the similar roles of ATP binding and hydrolysis in the eukaryotic and archaeal complexes make it likely that these Rpt subunits undergo similar coordinated conformational changes as does PAN in driving gate-opening, substrate unfolding and translocation. One obvious difference is that the 26S proteasome is a much more stable structure than the transitory PAN-20S complex, where association and gate-opening appear to be linked events, while in the 26S complex, association and gating can be uncoupled by single targeted point-mutations, since its different C-termini seem to play different roles in gating and association. Because of its homogeneity and ease of isolation and mutagenesis, PAN clearly offers major advantages for elucidating the gating mechanisms, and elsewhere we shall report the structural changes in the α-subunits that underlie gate-opening by PAN (Rabl et al, in preparation). Similar detailed understanding of this “key-in-a-lock” mechanism in the 26S proteasome will require precise knowledge about the sites of docking of each of the six ATPases’ C-termini in the core particle’s seven intersubunit pockets and the functional consequences of each binding event.
Experimental Procedures
Enzyme Assays
To measure gate-opening in the archaeal 20S, LFP (Mca-AKVYPYPME-Dpa(Dnp)-amide) in DMSO was used as a reporter substrate (Smith et al., 2005) at a final concentration 10μM, and for rabbit 20S, the indicated peptide in DMF was used at 100μM. The amount of DMSO or DMF did not exceed 1% of the total volume. To assay gate opening induced by WT PAN or the PAN variants the indicated PAN (1μg) and archaeal 20S (0.2μg) was incubated in the reaction buffer (50mM Tris pH 7.5, 1mM DTT and 10 mM MgCl2) with or without ATPγS (0.1mM) in a 100μl reaction volume. To monitor stimulation of gate opening by peptides for the archaeal or mammalian 20S proteasomes, the indicated peptides were added to the reaction buffer (100μl) at 45°C (archaeal 20S; 0.2 μg) or at 37°C (mammalian 20S; 0.1μg), and the rate of peptide hydrolysis was monitored.
Gate opening in rabbit muscle 20S proteasome either by the C-terminal peptides or by the 19S/PA700 complex was assayed similarly but with 3% glycerol in the reaction buffer and at 37°C with the indicated substrate. Assembly reactions for the mammalian 26S proteasome were performed as described (DeMartino, 2005). The 19S preparations contained a background activity that was not proteasomal (i.e. it cleaved nLPnLD-amc but not LLVY-amc and was not stimulated by ATP or ATPγS), and was thus subtracted out. Purified yeast proteasomes were subjected to non-denaturing PAGE and in-gel activity assay using LLVY-AMC as described (Elsasser et al., 2005). LFP hydrolysis was monitored at λex 380nm; λem 480nm (Smith et al., 2005) and amc hydrolysis at λex 380nm; λem 440nm. To follow unfolding of GFPssrA by carboxypeptidase treated PAN (1ug), its fluorescence was monitored in the reaction buffer at 45°C at λex 400nm and λem 510 nm. ATP hydrolysis by carboxypeptidase treated PAN was monitored as previously described (Ames, 1966). Experimental details regarding the tryptophan fluorescence experiments can be found in the Supplement.
Generation of PAN and yeast Rpt mutations
PAN mutation were constructed by PCR as described in the Supplement. Yeast mutations were constructed by integrative recombination and expressed from the appropriate endogenous chromosomal locus. While all mutant Yeast strains were viable most mutations cause various and highly complex phenotypes that will be presented elsewhere after further study. Details of strain constructions and genotypes of yeast strains used in this study are found in the supplement.
Supplementary Material
Acknowledgments
The authors are very grateful to Shuya Zhai for her assistance in preparing PAN, Mary Dethavong for her valuable assistance, Chris Hill for providing the 20S(K66A) plasmid, George DeMartino for providing purified PA700, and the members of the Goldberg lab for their useful discussions and comments on the manuscript. These studies were supported by a grant from the NIH (GM051923-09) to A.L.G., (GM43601) to D.F., the Fund for Innovation from the Elan Corp. to A.L.G., and. ALG is a Senior Fellow of the Ellison Foundation. DMS is supported by a Charles A. King Trust Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams J. Proteasome inhibitors in cancer therapy. Totowa, N.J: Humana Press; 2004. [Google Scholar]
- Ames BN. Assay of inorganic Phosphate, Total Phosphate and Phosphatases. Methods in Enzymology. 1966;8:115–118. [Google Scholar]
- Benaroudj N, Smith DM, Goldberg AL. What the archaeal PAN-proteasome complex and bacterial ATPdependent proteases can teach us about the 26S proteasome. Vol. 2. The Ubiquitin-Proteasome System; Wiley-VCH: 2005. [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. Embo J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annual Review of Biochemistry. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- DeMartino GN. Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods Enzymol. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. Embo J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Nobel committee tags ubiquitin for distinction. Neuron. 2005;45:339–344. doi: 10.1016/j.neuron.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Cascio P, Saric T, Rock KL. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/s0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure Of 20s Proteasome From Yeast At 2.4-Angstrom Resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol. 2003;35:606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Plenum Press; 1999. [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Ma CP, Willy PJ, Slaughter CA, DeMartino GN. PA28, an activator of the 20 S proteasome, is inactivated by proteolytic modification at its carboxyl terminus. J Biol Chem. 1993;268:22514–22519. [PubMed] [Google Scholar]
- Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- Ogura T, Tanaka K. Dissecting various ATP-dependent steps involved in proteasomal degradation. Mol Cell. 2003;11:3–5. doi: 10.1016/s1097-2765(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- Seong IS, Kang MS, Choi MK, Lee JW, Koh OJ, Wang J, Eom SH, Chung CH. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J Biol Chem. 2002;277:25976–25982. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening in the alpha-ring and translocation of unfolded proteins. Molecular Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. Journal of Structural Biology. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Clawson A, Realini C, Jensen CC, Knowlton JR, Hill CP, Rechsteiner M. Identification of an activation region in the proteasome activator REGalpha. Proc Natl Acad Sci U S A. 1998;95:2807–2811. doi: 10.1073/pnas.95.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. Journal of Biological Chemistry. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.