Abstract
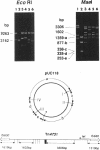
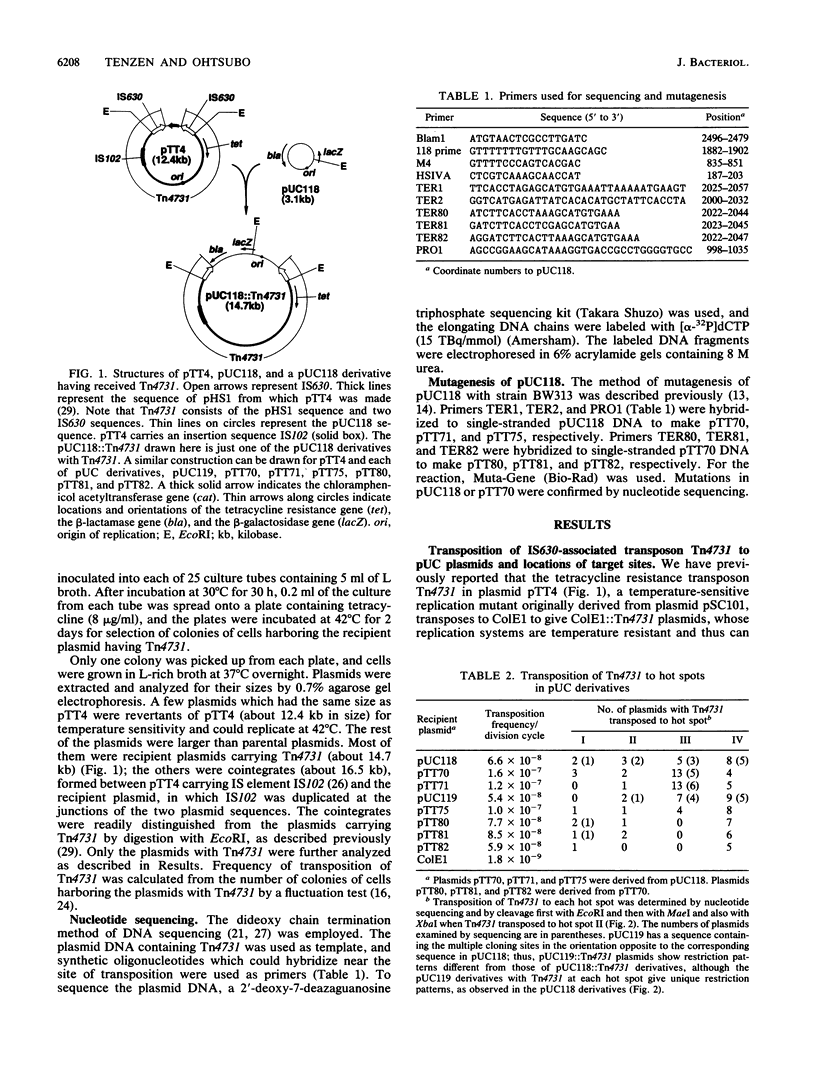
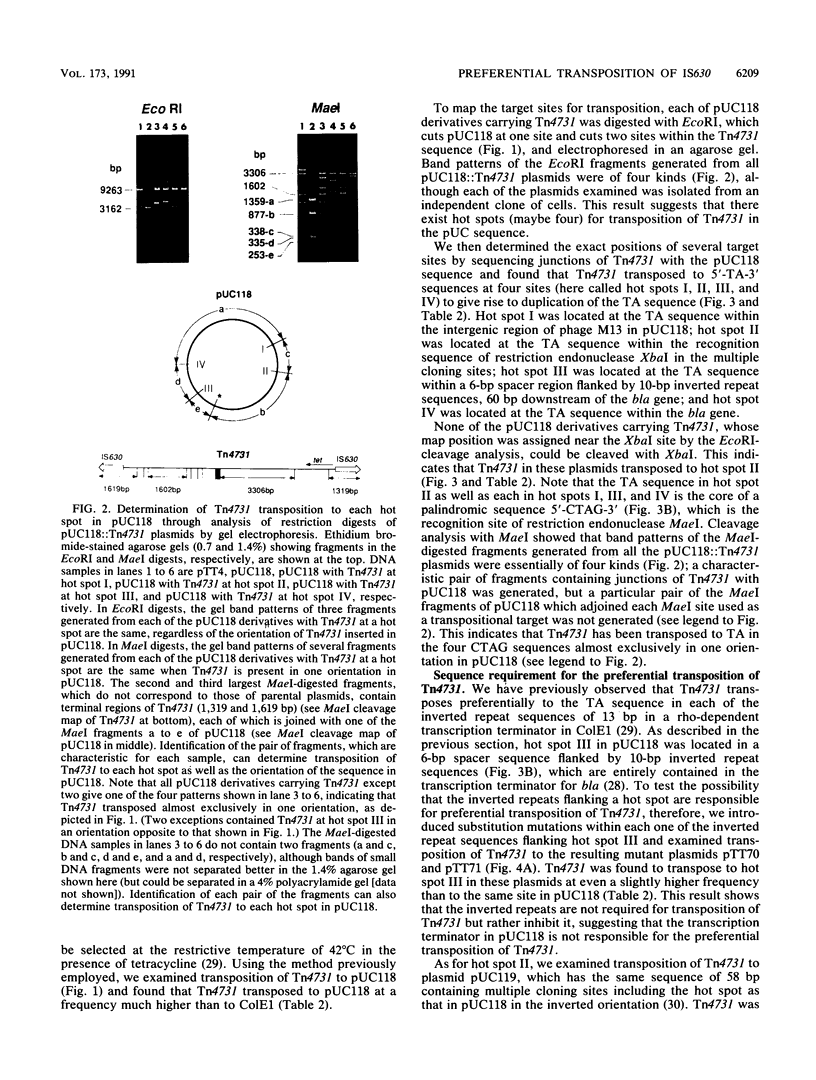
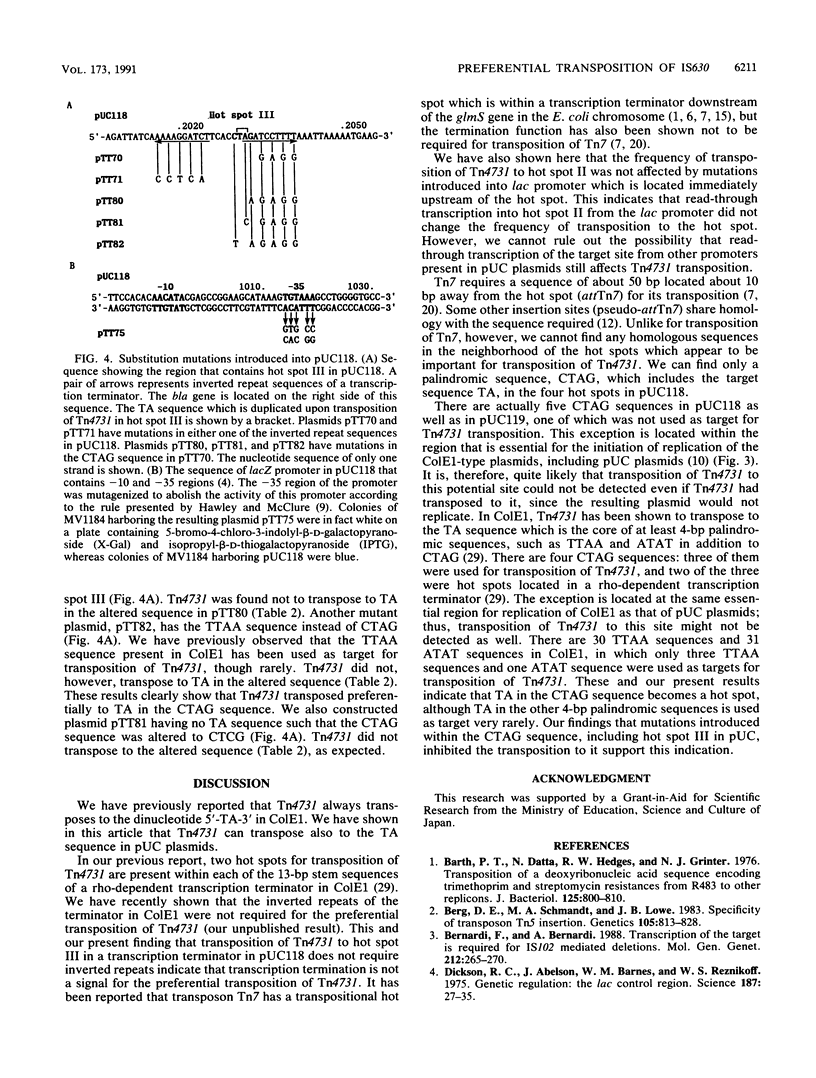
A composite transposon, Tn4731, associated with IS630 has been shown to transpose preferentially to 5'-TA-3' sequences that are located at two sites in a rho-dependent transcription terminator in plasmid ColE1 in Escherichia coli (T. Tenzen, S. Matsutani, and E. Ohtsubo, J. Bacteriol. 172:3830-3836, 1990). Here we demonstrated that Tn4731 preferentially transposes to TA sequences at four sites in plasmid pUC118 and its derivatives: the TA sequence (hot spot I) in the intergenic region of phage M13 within the pUC sequence, the TA sequence (hot spot II) in the XbaI site in multiple cloning sites of the lacZ coding region, the TA sequence (hot spot III) in a spacer region flanked by inverted repeat sequences of a transcription terminator located downstream of the bla gene, and the TA sequence (hot spot IV) in the middle of bla. Transposition of Tn4731 to hot spot III was found not to require the inverted repeats in the terminator. Transposition of Tn4731 to hot spot II, which is located immediately downstream of the lacZ promoter, was not affected by mutations introduced into the promoter. There appear to be no particular sequences important for transposition of Tn4731 around each of the hot spots, except a palindromic sequence, 5'-CTAG-3', that contains the target sequence. Mutations introduced within the CTAG sequence at a hot spot inhibited Tn4731 from transposing to it, indicating that the CTAG sequence is responsible for the preferential transposition of Tn4731.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Schmandt M. A., Lowe J. B. Specificity of transposon Tn5 insertion. Genetics. 1983 Dec;105(4):813–828. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F., Bernardi A. Transcription of the target is required for IS102 mediated deletions. Mol Gen Genet. 1988 May;212(2):265–270. doi: 10.1007/BF00334695. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Tybulewicz V. L., Walker J. E. Insertion of transposon Tn7 into the Escherichia coli glmS transcriptional terminator. Biochem J. 1986 Feb 15;234(1):111–117. doi: 10.1042/bj2340111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringauz E., Orle K. A., Waddell C. S., Craig N. L. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J Bacteriol. 1988 Jun;170(6):2832–2840. doi: 10.1128/jb.170.6.2832-2840.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaer R., Kühn S., Fritz H. J., Tillmann E., Saint-Girons I., Habermann P., Pfeifer D., Starlinger P. Studies on transposition mechanisms and specificity of IS4. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):215–224. doi: 10.1101/sqb.1981.045.01.034. [DOI] [PubMed] [Google Scholar]
- Kubo K. M., Craig N. L. Bacterial transposon Tn7 utilizes two different classes of target sites. J Bacteriol. 1990 May;172(5):2774–2778. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lichtenstein C., Brenner S. Unique insertion site of Tn7 in the E. coli chromosome. Nature. 1982 Jun 17;297(5867):601–603. doi: 10.1038/297601a0. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. A novel type of transposon generated by insertion element IS102 present in a pSC101 derivative. Cell. 1982 Aug;30(1):29–36. doi: 10.1016/0092-8674(82)90008-3. [DOI] [PubMed] [Google Scholar]
- Matsutani S., Ohtsubo H., Maeda Y., Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987 Aug 5;196(3):445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- McKown R. L., Orle K. A., Chen T., Craig N. L. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988 Jan;170(1):352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Murphy E., Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984 Jan 19;307(5948):292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Rosenbloom M., Schrempf H., Goebel W., Rosen J. Site specific recombination involved in the generation of small plasmids. Mol Gen Genet. 1978 Feb 16;159(2):131–141. doi: 10.1007/BF00270886. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Zenilman M., Ohtsubo E. Insertion element IS102 resides in plasmid pSC101. J Bacteriol. 1980 Oct;144(1):131–140. doi: 10.1128/jb.144.1.131-140.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res. 1985 Sep 11;13(17):6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Matsutani S., Ohtsubo E. Site-specific transposition of insertion sequence IS630. J Bacteriol. 1990 Jul;172(7):3830–3836. doi: 10.1128/jb.172.7.3830-3836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y., Ohtsubo H., Ohtsubo E. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol. 1987 Feb;169(2):619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]