Abstract
A gene (lat) encoding lysine 6-aminotransferase was found upstream of the pcbAB (encoding alpha-aminoadipylcysteinyl-valine synthetase) and pcbC (encoding isopenicillin N synthase) genes in the cluster of early cephamycin biosynthetic genes in Nocardia lactamdurans. The lat gene was separated by a small intergenic region of 64 bp from the 5' end of the pcbAB gene. The lat gene contained an open reading frame of 1,353 nucleotides (71.4% G + C) encoding a protein of 450 amino acids with a deduced molecular mass of 48,811 Da. Expression of DNA fragments carrying the lat gene in Streptomyces lividans led to a high lysine 6-aminotransferase activity which was absent from untransformed S. lividans. The enzyme was partially purified from S. lividans(pULBS8) and showed a molecular mass of 52,800 Da as calculated by Sephadex gel filtration and polyacrylamide gel electrophoresis. DNA sequences which hybridized strongly with the lat gene of N. lactamdurans were found in four cephamycin-producing Streptomyces species but not in four other actinomycetes which are not known to produce beta-lactams, suggesting that the gene is specific for beta-lactam biosynthesis and is not involved in general lysine catabolism. The protein encoded by the lat gene showed similarity to ornithine-5-aminotransferases and N-acetylornithine-5-aminotransferases and contained a pyridoxal phosphate-binding consensus amino acid sequence around Lys-300 of the protein. The evolutionary implications of the lat gene as a true beta-lactam biosynthetic gene are discussed.
Full text
PDF

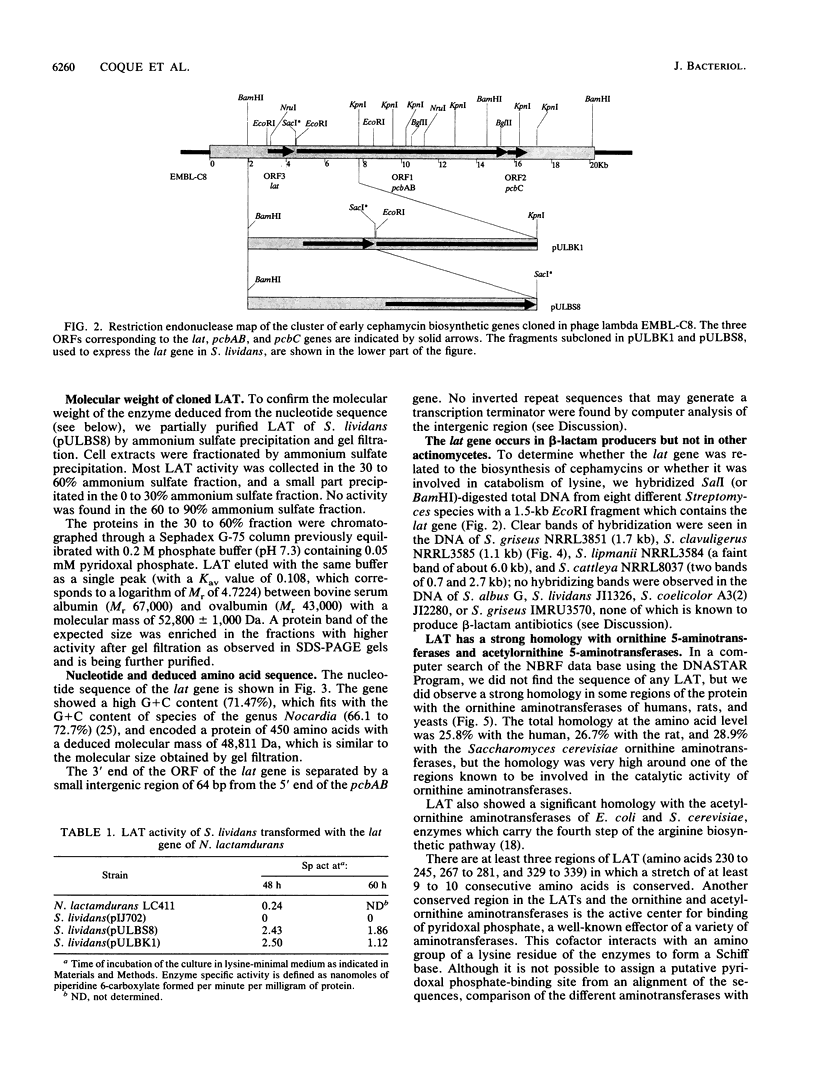




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. E., Bird J. W., Field R. A., O'Callaghan N. M., Schofield C. J., Willis A. C. Isolation and partial characterisation of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. Evidence for the presence of phosphopantothenate in ACV synthetase. J Antibiot (Tokyo) 1991 Feb;44(2):241–248. doi: 10.7164/antibiotics.44.241. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee J. K. alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes. Crit Rev Microbiol. 1985;12(2):131–151. doi: 10.3109/10408418509104427. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cohen G., Shiffman D., Mevarech M., Aharonowitz Y. Microbial isopenicillin N synthase genes: structure, function, diversity and evolution. Trends Biotechnol. 1990 Apr;8(4):105–111. doi: 10.1016/0167-7799(90)90148-q. [DOI] [PubMed] [Google Scholar]
- Coque J. J., Martín J. F., Calzada J. G., Liras P. The cephamycin biosynthetic genes pcbAB, encoding a large multidomain peptide synthetase, and pcbC of Nocardia lactamdurans are clustered together in an organization different from the same genes in Acremonium chrysogenum and Penicillium chrysogenum. Mol Microbiol. 1991 May;5(5):1125–1133. doi: 10.1111/j.1365-2958.1991.tb01885.x. [DOI] [PubMed] [Google Scholar]
- Der Garabedian P. A., Vermeersch J. J. Lysine degradation in Candida. Characterization and probable role of L-norleucine-leucine, 4-aminobutyrate and delta-aminovalerate:2-oxoglutarate aminotransferases. Biochimie. 1989 Apr;71(4):497–503. doi: 10.1016/0300-9084(89)90180-6. [DOI] [PubMed] [Google Scholar]
- Drew S. W., Demain A. L. Effect of primary metabolites on secondary metabolism. Annu Rev Microbiol. 1977;31:343–356. doi: 10.1146/annurev.mi.31.100177.002015. [DOI] [PubMed] [Google Scholar]
- Díez B., Gutiérrez S., Barredo J. L., van Solingen P., van der Voort L. H., Martín J. F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990 Sep 25;265(27):16358–16365. [PubMed] [Google Scholar]
- Fothergill J. C., Guest J. R. Catabolism of L-lysine by Pseudomonas aeruginosa. J Gen Microbiol. 1977 Mar;99(1):139–155. doi: 10.1099/00221287-99-1-139. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- García-Domínguez M., Liras P., Martín J. F. Cloning and characterization of the isopenicillin N synthase gene of Streptomyces griseus NRRL 3851 and studies of expression and complementation of the cephamycin pathway in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1991 Jan;35(1):44–52. doi: 10.1128/aac.35.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J. A., Naharro G., Villanueva J. R., Martín J. F. Characterization and regulation of p-aminobenzoic acid synthase from Streptomyces griseus. J Gen Microbiol. 1985 Jun;131(6):1279–1287. doi: 10.1099/00221287-131-6-1279. [DOI] [PubMed] [Google Scholar]
- Gutiérrez S., Díez B., Montenegro E., Martín J. F. Characterization of the Cephalosporium acremonium pcbAB gene encoding alpha-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol. 1991 Apr;173(7):2354–2365. doi: 10.1128/jb.173.7.2354-2365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg H., Boyen A., Crabeel M., Glansdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferase: evolutionary relationship with ornithine aminotransferase. Gene. 1990 May 31;90(1):69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kern B. A., Hendlin D., Inamine E. L-lysine epsilon-aminotransferase involved in cephamycin C synthesis in Streptomyces lactamdurans. Antimicrob Agents Chemother. 1980 Apr;17(4):679–685. doi: 10.1128/aac.17.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel J. J., Winston M. K., Bhattacharjee J. K. Role of L-lysine-alpha-ketoglutarate aminotransferase in catabolism of lysine as a nitrogen source for Rhodotorula glutinis. J Bacteriol. 1983 Jul;155(1):417–419. doi: 10.1128/jb.155.1.417-419.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch J. F., Eichele G., Ford G. C., Vincent M. G., Jansonius J. N., Gehring H., Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984 Apr 15;174(3):497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- Kovacevic S., Miller J. R. Cloning and sequencing of the beta-lactam hydroxylase gene (cefF) from Streptomyces clavuligerus: gene duplication may have led to separate hydroxylase and expandase activities in the actinomycetes. J Bacteriol. 1991 Jan;173(1):398–400. doi: 10.1128/jb.173.1.398-400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCabe A. P., Riach M. B., Unkles S. E., Kinghorn J. R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990 Jan;9(1):279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri K., Stuttard C., Vining L. C. Cloning and location of a gene governing lysine epsilon-aminotransferase, an enzyme initiating beta-lactam biosynthesis in Streptomyces spp. J Bacteriol. 1991 Feb;173(3):985–988. doi: 10.1128/jb.173.3.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri K., Stuttard C., Vining L. C. Lysine catabolism in Streptomyces spp. is primarily through cadaverine: beta-lactam producers also make alpha-aminoadipate. J Bacteriol. 1989 Jan;171(1):299–302. doi: 10.1128/jb.171.1.299-302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín J. F., Liras P. Enzymes involved in penicillin, cephalosporin and cephamycin biosynthesis. Adv Biochem Eng Biotechnol. 1989;39:153–187. doi: 10.1007/BFb0051954. [DOI] [PubMed] [Google Scholar]
- Martín M. F., Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Sequence of the precursor to rat ornithine aminotransferase deduced from a cDNA clone. J Biol Chem. 1985 Oct 25;260(24):12993–12997. [PubMed] [Google Scholar]
- Navarrete R. M., Vara J. A., Hutchinson C. R. Purification of an inducible L-valine dehydrogenase of Streptomyces coelicolor A3(2). J Gen Microbiol. 1990 Feb;136(2):273–281. doi: 10.1099/00221287-136-2-273. [DOI] [PubMed] [Google Scholar]
- Ramón D., Carramolino L., Patiño C., Sánchez F., Peñalva M. A. Cloning and characterization of the isopenicillin N synthetase gene mediating the formation of the beta-lactam ring in Aspergillus nidulans. Gene. 1987;57(2-3):171–181. doi: 10.1016/0378-1119(87)90120-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. J., Burnham M. K., Bull J. H., Hodgson J. E., Ward J. M., Browne P., Brown J., Barton B., Earl A. J., Turner G. Beta-lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990 Mar;9(3):741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda K., Misono H., Yamamoto T. L-Lysine:alpha-ketoglutarate aminotransferase. I. Identification of a product, delta-1-piperideine-6-carboxylic acid. Biochemistry. 1968 Nov;7(11):4102–4109. doi: 10.1021/bi00851a045. [DOI] [PubMed] [Google Scholar]
- van Liempt H., von Döhren H., Kleinkauf H. delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans. The first enzyme in penicillin biosynthesis is a multifunctional peptide synthetase. J Biol Chem. 1989 Mar 5;264(7):3680–3684. [PubMed] [Google Scholar]




