Abstract
Talin is an actin-binding protein involved in integrin-mediated cell adhesion and spreading. The C-terminal 197 amino acids of vertebrate talin are 45% similar to the C-terminal residues of Sla2, a yeast protein implicated in polarized assembly of the yeast actin cytoskeleton. Talin is also homologous in this region to nematode talin, cellular slime mold filopodin, and an Sla2 homolog from nematode. Analysis of the conserved C-terminal sequences of these five proteins with block maker reveals a series of four blocks, which we name the I/LWEQ module after the conserved initial residues in each block. Experiments presented here show that the conserved protein domain represented by the I/LWEQ module competes quantitatively with native talin for binding to F-actin in vitro. Furthermore, the corresponding domain of Sla2 binds to both yeast and vertebrate F-actin in vitro. Mutation of one of the conserved residues in the fourth conserved block abolishes the interaction of the Sla2 I/LWEQ module with F-actin. These results establish the location of an F-actin binding domain in native talin, demonstrate that direct interaction of Sla2 with actin is a possible basis for its effect on the actin cytoskeleton in vivo, and define the I/LWEQ consensus as a new actin-binding motif.
Talin is a modular protein (1, 2) found in focal adhesions (3), multiprotein assemblies mediating interactions between the actin cytoskeleton of cultured cells and the extracellular environment (4). Perturbation of talin in vivo indicates that talin functions in cell adhesion and spreading (5–7). Experiments with purified proteins demonstrate that talin interacts with several focal adhesion components, including acidic phospholipids (8), actin (9–11), vinculin (12), β1 integrin (13), β3 integrin (14), and focal adhesion kinase (FAK) (15). Talin has a calculated monomer mass of 269 kDa and is a dimer in its actin-binding form (16, 17) and at protein concentrations greater than 1 μM (16). Calpain cleaves talin into two fragments that have different activities in vitro. The 190-kDa, C-terminal fragment binds to G and F-actin (2, 9) and to vinculin (18), and nucleates actin polymerization (17). Recent studies with glutathione S-transferase (GST) fusion proteins provide evidence for two, nonoverlapping F-actin binding sequences in the 190-kDa talin fragment (19), and one F-actin binding sequence from the N-terminal, 47-kDa region. In other studies, the 47-kDa proteolytic fragment did not bind to F-actin or nucleate actin polymerization (2, 17). The 47-kDa fragment contains all of the acidic phospholipid binding activity of intact talin (2), which has led to the hypothesis that the N terminus of talin mediates talin-membrane interactions in vivo.
Several proteins have been identified that are similar to one or more limited regions of talin, suggesting that the specific functions associated with individual talin domains are dispersed among proteins having cellular roles distinct from those of talin. These proteins appear to fall into three groups. The first of these is the erythrocyte band 4.1 superfamily. Talin is a member of this group based on similarity between the 47-kDa fragment of talin and the N-terminal region of erythrocyte band 4.1 (1). The 4.1 superfamily also includes the related cytoskeletal proteins ezrin, moesin, and radixin, the tumor suppressor schwannomin (NF-2), and several protein tyrosine phosphatases (20). All of these proteins are related by a homologous 4.1-like domain. All members of the 4.1 superfamily are known, or potential, membrane-associated proteins.
A second group of proteins contains members that are similar to vertebrate talin at their N-terminal 4.1 region and at their C termini. One such talin homolog has been identified in Dictyostelium (21). This 2,491-residue protein, filopodin, accumulates in response to chemoattractant at the leading edge of motile cells, where F-actin is also enriched. Other than at their N and C termini, talin and filopodin are not very similar. In contrast to filopodin, an apparently bona fide talin has been identified in Caenorhabditis elegans (22). This 2,553-residue talin is 59% similar to mouse talin over its entire length, with greatest similarity at its N terminus, including the band 4.1 region, and at its C terminus.
A third group of proteins is homologous to talin only at the C-terminal 200 residues. This group is represented by Sla2, a 968-residue yeast protein that is required for polarized assembly of the actin cytoskeleton (23). SLA2 is allelic with END4, whose product is required for endocytosis (24), and with MOP2, which is necessary for the proper plasma membrane localization of the H+-ATPase encoded by PMA1 (25). Talin and Sla2 (End4/Mop2) also share their similar C terminus with a putative Sla2 protein from C. elegans (26).
Because Sla2 is required for nucleated assembly of actin in a permeabilized yeast cell model (27), and because talin both nucleates actin polymerization and binds to F-actin in vitro (9, 10, 17), we hypothesized that Sla2 might interact directly with actin and that the actin remodeling activities of talin and Sla2 reside in their conserved C-terminal domain. To test this idea, we constructed two GST-fusion proteins incorporating the C-terminal 197 residues of mouse talin and the homologous residues of yeast Sla2 and evaluated their potential to interact with vertebrate and yeast actin.
MATERIALS AND METHODS
Protein Preparation.
Rabbit skeletal muscle actin was purified as described (28), with an additional gel filtration step (29). Pyrene-labeled actin was produced by labeling F-actin with pyrene iodoacetamide (30, 31). Yeast actin (32) and chicken gizzard talin (33) were purified as described.
Protein Sequence Analysis.
The C-terminal sequences of talin and its putative homologs were aligned and blocks 1–4 of the I/LWEQ module identified using block maker (34) [http://www.blocks.fhcrc.org/].
GST Fusion Proteins.
The mouse talin GST fusion protein construct (GST-Tn.2345–2541) was prepared by amplifying the coding region extending from I2345 to the stop codon, using a mouse talin cDNA as the template, and subcloning this PCR fragment into pCR 2.1 (Invitrogen). The following primers were used: 5′-ATCCTAGAAGCTGCC-3′ (7192–7206; Start=I2345); 5′-TTAGTGCTCGTCTCG-3′ [7785–7771; numbering corresponds to that in Rees et al. (1)]. The EcoRI insert-containing fragment from pCR2.1 was then subcloned into EcoRI-digested pGEX-2T (Pharmacia). The Sla2 GST fusion protein construct (GST-Sla2.771–968) was prepared similarly, except that yeast genomic DNA (strain SM1060) was used as the template. The following primers were used for the Sla2 construct: 5′-CCATTGTTGTCATTGGC-3′ (Chr XIV: 190360–190376; Start=P771); 5′-GATCAATCATCATCCTGG-3′ (Chr XIV: 190958–190944; numbering from the Saccharomyces Genome Database). The fusion proteins were purified using glutathione-agarose as previously described (35).
F-Actin Cosedimentation.
Cosedimentation of mouse talin, GST-Tn.2345–2541, and GST-Sla2.772–968 with F-actin was measured essentially as described in Schmidt et al. (11) by mixing G-actin (in buffer G: 2 mM Tris, pH 7.5, 9490.2 mM CaCl2/0.2 mM ATP/0.2 mM dithiothreitol) with talin or one of the GST fusion proteins and then initiating actin polymerization by adding MgCl2 to 2 mM and KCl to 50 mM (50 μl total volume). After polymerization was complete (60 min, 22°C) the mixture was centrifuged at 150,000 g (Beckman Airfuge) for 20 min and the supernatant and pellet were separated for subsequent analysis on SDS/PAGE.
F-Actin Bundling.
Actin (3 μM) was polymerized in the presence of GST, GST-Tn.2345–2541, or GST-Sla2.771–968 (3 μM each). The resulting F-actin preparations were adsorbed to grids, negatively stained with uranyl formate, and visualized on a Zeiss model 10A electron microscope.
Nucleation of Actin Polymerization.
G-actin (6 μM, 54% pyrene-labeled) was polymerized as outlined above in the presence of talin, GST-Tn, GST-Sla2, or GST (2 μM each), except for the addition of 30 mM NaCl and a final Tris concentration of 7 mM. Polymerization was monitored by the increase in pyrene fluorescence (Perkin–Elmer model LS 50 B; excitation: 365 nm; emission 407 nm). Gelsolin (2 nM) was used as a positive control for nucleation.
RESULTS
At the start of this study, block maker (34) was used to aid in identification of the most conserved patterns of amino acids in the conserved C termini of the four proteins then identified as talin homologs (1, 21, 23, 26). block maker uses automated versions of motifs (36) and gibbs (37) in conjunction with motomat (38) for identification of the best set of conserved amino acid patterns (motifs) in homologous protein sequences. The motifs algorithm with the parameters s, r, and d (36) automatically fixed at 4, 0, and 17, respectively, found blocks 1–4 (Fig. 1) when queried with all four sequences. Recently, the sequence of C. elegans talin was reported (22), and when it was included in the block maker analysis, motifs no longer found block 1 because the values for s, r, and d automatically change to 5, 0, and 17. This means that in the block 1 region there is no longer an amino acid triplet pattern of the form aa1-d1-aa2-d2-aa3 in which the amino acids at positions 1, 2, and 3 are identical across the five sequences, as specified by the value for s. In Fig. 1 the columns of amino acids that are identical between the five aligned proteins are shaded. Positions that are identical in at least four out of the five sequences are indicated by a dot above the column. The gibbs algorithm consistently found blocks 2 and 4.
Figure 1.
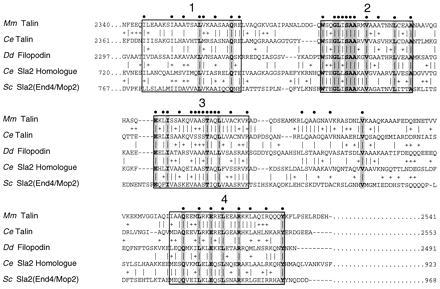
I/LWEQ module sequence alignment. Alignment of the conserved C-terminal domain common to mouse (Mm) and C. elegans (Ce) talin, the Dictyostelium (Dd) talin homolog filopodin, the putative C. elegans Sla2 homolog, and S. cerevisiae (Sc) Sla2. The four conserved blocks, originally identified using the block maker program (34), are boxed; identities between all five sequences are highlighted by shading in the column, conservative substitutions are indicated by +. The most conserved positions (at least four out of five residues identical) are marked by a solid circle above the column. Block 1 contains 17% identities and 48% similarites; the values for blocks 2–4 are 63%/80%, 57%/83%, and 46%/66%, respectively.
Based on the patterns found in the initial four proteins we designed fusion proteins starting at the beginning of block 1. We have named the conserved C-terminal set of motifs the I/LWEQ module, after the conserved initial residues of blocks 1–4 (Fig. 1). To test the hypothesis that the I/LWEQ module interacts with actin, we constructed GST-Tn.2345–2541 (GST-Tn), which begins with the initial residue of block 1 of mouse talin and extends to the C terminus, and GST-Sla2.771–968 (GST-Sla2), which begins with Pro-771, immediately N-terminal to block 1 of yeast Sla2, and extends to the C terminus. Thus both GST-fusions include all four conserved blocks from their respective proteins. We then examined these fusion proteins for interaction with vertebrate and yeast actin in vitro.
Binding of talin, GST-Tn and GST-Sla2 to F-actin was assayed by measuring cosedimention with vertebrate F-actin (Fig. 2). With increasing actin concentration, native talin, GST-Tn, and GST-Sla2 are depleted from the residual supernatant following ultracentrifugation, and this depletion is accompanied by an increase of talin or each fusion protein in the corresponding pellet fractions (Fig. 2A). Cosedimentation of GST-Tn and GST-Sla2 with F-actin could not be measured directly by densitometry of the fusion protein pellet fractions because the large amount of sedimented actin obscures the fusion proteins; therefore we used supernatant depletion of the fusion proteins as a measure of cosedimentation. Immunoblots of the fusion protein pellet fractions with anti-GST showed that depletion of GST-Tn and GST-Sla2 from their respective supernatants was accompanied by enrichment in their respective F-actin pellet fractions (Fig. 2A). With native talin and GST, where it was possible to measure directly the amount of protein in each supernatant and pellet sample from densitometry of Coomassie blue stained gels, protein depleted from each supernatant fraction could be accounted for quantitatively in the corresponding pellet. GST did not bind to F-actin, and the GST-fusion proteins did not sediment in the absence of F-actin (Fig. 2A).
Figure 2.
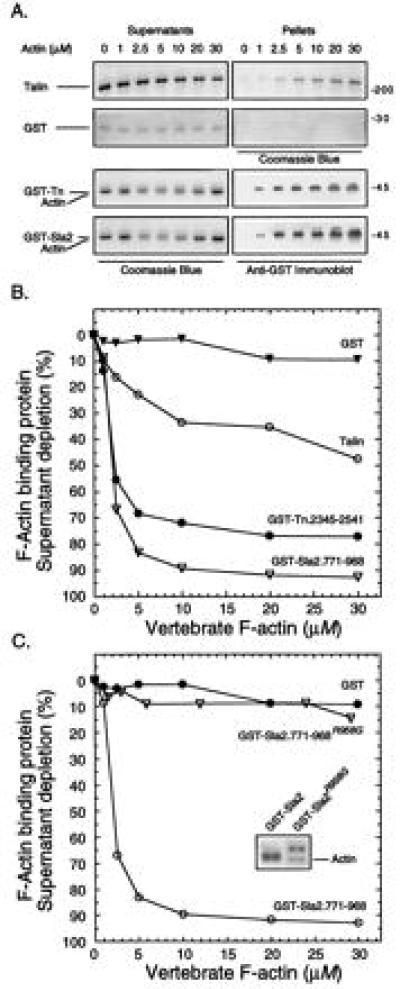
F-actin cosedimentation: vertebrate actin. (A) SDS/PAGE. Rabbit skeletal muscle actin (0–30 μM) was mixed with either native mouse talin (2.5 μM), GST-Tn (2.0 μM), GST-Sla2 (2.0 μM), or GST (2.0 μM), and F-actin cosedimentation analyzed. All of the supernatant panels and the talin and GST pellet panels were visualized with Coomassie blue; the fusion protein pellet fractions were detected by immunoblotting (see text). (B) Quantitative analysis. Supernatant depletion, as a measure of cosedimentation, of talin (○), GST-Tn (•), GST-Sla2 (▿), and GST (▾) was determined densitometrically using the gels shown in A. (C) Quantitative analysis: GST-Sla2 mutant. Supernatant depletion was performed as in A on GST-Sla2 (○), the mutant GST-Sla2R958G(▿), and GST (•). The shift in electrophoretic mobility of the Sla2 mutant is shown (Inset).
A quantitative analysis of the gels illustrated in Fig. 2A is shown in Fig. 2B. Both GST-fusion proteins bound to F-actin to a greater extent than native talin, with GST-Sla2 binding to a greater extent than GST-Tn. The actin concentration required for half-maximal binding of fusion protein was ≈3 μM for GST-Tn and GST-Sla2 and ≈5 μM for native talin. In this particular experiment, ≈45% of the talin bound to F-actin at the saturating actin concentration (30 μM), which is within the range we have found in several other experiments using different talin and actin preparations.
Other investigators have also observed that only a portion of purified native talin cosediments with actin. In fact, in the earliest studies (3, 12) none of the purified chicken gizzard talin cosedimented with actin. Subsequently other investigators reported that talin does bind actin (9–11). Evidence suggests that the amount of active talin depends on the purification protocol (9) and on the conditions of ionic strength and pH used in the binding assay (11). Our preparations contain 45–60% active talin that was not significantly affected by pH (in the range between 6.6–7.4), or length of incubation with actin (up to 24 hr) in the cosedimentation assay. All talin molecules in our preparation have an intact C terminus as assessed by quantitative immunoprecipitation with an antibody raised to the C-terminal 21 amino acids of murine talin (data not shown). Thus the molecular basis for functional heterogeneity in our talin preparations is unknown.
One of the GST-Sla2 clones generated by PCR yielded a fusion protein that failed to bind to F-actin (Fig. 2C). The DNA sequence of this clone showed a single base change resulting in an R to G substitution at position 958 in Sla2. This mutant had a slightly retarded migration on SDS gels compared with the wild-type fusion protein (Fig. 2C Inset), suggesting that the mutation affects a structural fold on which the amino acids contacting actin are arrayed. This function-blocking mutation at a conserved residue of the I/LWEQ domain provides preliminary support for the hypothesis that the conserved residues of the I/LWEQ module are responsible for the actin-binding activity of the module.
Yeast and vertebrate actin are 87% identical and 94% similar, which is substantially less homologous than the six vertebrate actins are to each other. To test the assumption of functional homology, the I/LWEQ modules of mouse talin and yeast Sla2 were tested for binding to yeast F-actin. The results show that mouse talin, GST-Tn and GST-Sla2 also interact with yeast F-actin (Fig. 3). Once again both GST-Tn and GST-Sla2 bound to F-actin to a greater extent than native talin, with GST-Sla2 binding to the greatest extent. GST alone did not bind to yeast F-actin. Therefore, the I/LWEQ module from both a vertebrate and a yeast protein is able to mediate an interaction in vitro with F-actin from either source.
Figure 3.
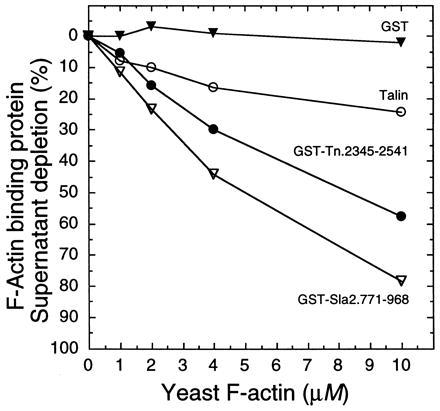
F-actin cosedimentation: yeast actin. Cosedimentation of talin (○), GST-Tn (•), GST-Sla2 (▿), and GST (▾) with yeast F-actin was measured as described in Fig. 2 except for the following changes in protein concentrations: yeast actin (0–10 μM); talin, GST-Tn, GST-Sla2, and GST (2.0 μM each).
To evaluate the extent to which the I/LWEQ module accounts for the interaction between native talin and F-actin, GST-Tn was tested for its ability to compete with talin for binding to F-actin (Fig. 4). In this experiment, ≈55% of native talin bound to vertebrate F-actin at a 10:1 molar ratio of actin to talin. Increasing the GST-Tn concentration from zero to 20 μM completely displaced the talin from F-actin (GST alone had no effect; data not shown). This result indicates that the I/LWEQ module is responsible for all of talin’s F-actin binding capacity, as measured in vitro by cosedimentation. This finding suggests that the additional F-actin binding sites recently identified in talin using GST-fusion proteins (19) may be inaccessible in the purified molecule and further, raises the interesting possibility that the full actin-binding activity of talin may be conformationally regulated, as recently demonstrated for vinculin (39).
Figure 4.
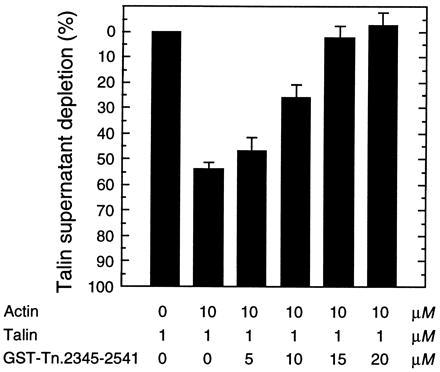
Competition of GST-Tn.2345–2541 for talin F-actin binding activity. Rabbit actin (10 μM) and talin (1 μM) were mixed in the presence of increasing amounts of GST-Tn (0–20 μM), and actin polymerization, centrifugation, SDS/PAGE, and densitometry were performed as previously described (Fig. 2). Values reported are averages of three separate experiments. GST (0–20 μM) had no effect on talin binding to F-actin (not shown).
Talin nucleates actin polymerization at the relatively high molar ratio of 1:3 (talin/actin) (2, 10). When tested for similar activity, neither GST-Tn nor GST-Sla2 nucleated actin polymerization. Native talin at a ratio of 1:3 (talin/actin) did shorten the lag phase associated with actin polymerization (Fig. 5). By contrast, gelsolin is a much more efficient nucleator of actin polymerization (Fig. 5).
Figure 5.

F-actin bundling. Actin (3 μM) was polymerized in the presence of (A) talin, (B) GST, (C) GST-Tn, or (D) GST-Sla2 (3 μM each), and the resulting F-actin preparations were negatively stained with uranyl formate and visualized by electron microscopy. (Bar = 10 μM).
Native talin induces the assembly of thick bundles of actin filaments (Fig. 6A), which are similar to those observed in other studies (40), whereas GST-Tn and GST-Sla2 aggregate actin filaments into small bundles (Fig. 6 C and D). GST alone had no discernible effect on filament morphology (Fig. 6B). Talin is a homodimer when it binds to actin (16), GST is a dimer (41, 42), and GST and our fusion proteins migrate as dimers by gel filtration analysis in physiological saline (data not shown). Thus differences in the properties of talin and the I/LWEQ domain fusion proteins do not reflect differences in monomer vs. dimer configurations.
Figure 6.
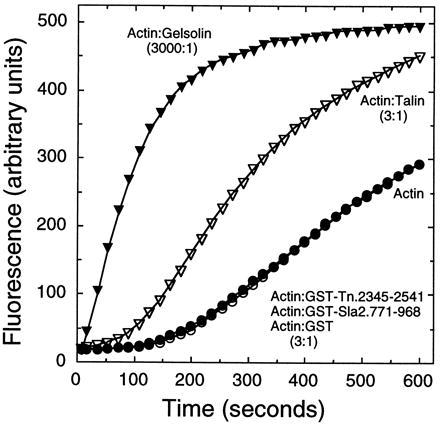
Nucleation of F-actin polymerization. Actin (6 μM, 54% pyrene-labeled) was polymerized alone (○) or in the presence of talin (2 μM, ▿), GST-Tn, GST-Sla2, or GST (2 μM each (•), or gelsolin (2 nM, ▾) and polymerization was monitored by the increase in pyrene fluorescence.
The five proteins that possess an I/LWEQ module are members of a talin superfamily that includes the band 4.1 superfamily and the Sla2 family. Representative members of these families are diagrammed to relative scale in Fig. 7. The I/LWEQ actin-binding module is indicated by the four shaded C-terminal blocks. The presence of the I/LWEQ module groups the talins and Sla2 proteins. Although the I/LWEQ module is not found in the ezrin, moesin, and radixin family, some ezrin, moesin, and radixin proteins do have a C-terminal actin binding domain (43). The presence of an N-terminal band 4.1 region groups talins, filopodin, and members of the ezrin, moesin, and radixin family (represented by ezrin). The conserved N-terminal blocks in yeast Sla2 and its C. elegans homolog are distinct from those of the band 4.1 superfamily.
Figure 7.
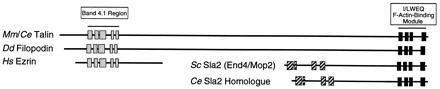
I/LWEQ actin-binding module superfamily. The five proteins containing the I/LWEQ module (mouse and C. elegans talin are represented together) are diagrammed to relative scale. (░⃞) Conserved sequence blocks identified using block maker (34). See text for discussion.
Fig. 7 also shows that the I/LWEQ-containing proteins can be grouped into at least two classes; one consisting of Mus musculus (vertebrate) and C. elegans (invertebrate) talin plus D. discoideum filopodin and another composed of Saccharomyces cerevisiae Sla2 and its putative C. elegans homolog. Filopodin is clearly related to talin by virtue of its band 4.1 region and I/LWEQ module, but it is not otherwise talin. Thus, filopodin may represent a third class of I/LWEQ-containing proteins, with its ultimate classification depending on further molecular, cellular, and genetic characterization.
DISCUSSION
We have shown that the conserved C-terminal domains of yeast Sla2 and mammalian talin represent a novel actin binding element. This conserved sequence element, which we call the I/LWEQ module, binds to F-actin and bundles actin filaments. The I/LWEQ module is unrelated to the F-actin binding motif found in the α-actinin family that includes ABP 120, fimbrin, dystrophin, filamin, and spectrin (44).
Conservation of sequence and F-actin binding function of the I/LWEQ domain between yeast and vertebrate proteins allows some new predictions. Although the direct interaction of talin with actin in vitro is well established, such information on the other I/LWEQ proteins is lacking. Filopodin was identified in a screen using a panel of monoclonal antibodies raised against Dictyostelium proteins that had been enriched on an F-actin affinity matrix, and the protein accumulates at the tips of filopodia, cell surface extensions that contain a core of bundled F-actin filaments (21). Although it is not known whether filopodin interacts directly with F-actin, the conserved function of the I/LWEQ module from mouse talin and yeast Sla2 strongly implies that the I/LWEQ module of filopodin mediates a filopodin/F-actin interaction in Dictyostelium. The finding that GST-Tn. 2345–2541 and GST-Sla2.771–968 bundle F-actin filaments in vitro suggests that filopodin, through its I/LWEQ module, bundles actin filaments in Dictyostelium filopodia.
SLA2 was identified in a synthetic lethal screen as being required along with ABP1 for polarized assembly of the actin cytoskeleton (23). SLA2 is also necessary for polarized nucleation of cortical actin filament assembly in permeabilized yeast cells (27). Although the I/LWEQ module of Sla2 mediates an interaction with vertebrate and yeast F-actin, GST-Sla2.771–968 did not nucleate (vertebrate) actin polymerization in vitro. However, this negative result may not reflect the properties of intact Sla2 because the in vitro nucleation activity of talin was not reproduced by GST-Tn.2345–2541.
Our data show that the I/LWEQ module competes quantitatively for the interaction of talin with F-actin in cosedimentation assays, but does not reproduce all of the effects of native talin on actin assembly and structure. Reasons for the differences between intact talin and the I/LWEQ module remain to be defined, but probably reflect the additional N-terminal actin-binding sequences in talin (19), or a requirement for a particular arrangement (e.g., antiparallel) of the I/LWEQ domains in the GST fusion protein dimer. Because all of the F-actin binding activity of native talin is competed by the I/LWEQ domain, we hypothesize that the additional actin-binding sequences that have been described by fusion protein analysis (19) are cryptic in native talin and require some type of regulated exposure for function.
The observation that yeast genes END4 (24) and MOP2 (25), are allelic with SLA2 (Saccharomyces Genome Database) raises some interesting possibilities. Several lines of evidence indicate that intracellular vesicle trafficking in yeast is polarized and associated with the actin cytoskeleton (45, 46). Identification of the Sla2(End4) I/LWEQ module as an F-actin binding domain provides evidence for a direct molecular connection between the vesicle traffic associated with fluid-phase and receptor-mediated endocytosis in yeast and the actin cytoskeleton. That Sla2(Mop2) is also required for the proper accumulation and/or maintenance of Pma1 indicates that the organization of some plasma membrane proteins may be connected via Sla2/End4/Mop2 to the actin cytoskeleton.
In summary, characterization of the conserved C-terminal I/LWEQ module of talin, and Sla2 as a novel actin-binding element has identified a molecular function that may connect several ostensibly diverse cellular processes, including focal adhesion function, yeast cortical actin organization, Dictyostelium locomotion, intracellular vesicle traffic, and plasma membrane organization, with the actin-based cytoskeleton. The involvement of these I/LWEQ-containing proteins in several different cellular processes in experimental systems that are amenable to molecular, cellular, and genetic approaches should facilitate the elucidation of how the domain functions in vivo.
Acknowledgments
We thank P. A. Rubenstein, A. E. M. Adams, and E. De La Cruz for advice on yeast actin purification; R. P. Johnson for help with protein purification and for reading the manuscript; P. Steimle for reading the manuscript; S. Michaelis for comments on the manuscript and K. Fujimura-Kamada for yeast genomic DNA; R. O. Hynes for mouse talin cDNA; and P. Pedersen and T. Golden for use of the fluorimeter. This work was supported by National Institutes of Health Grant GM41605 and a grant from the Muscular Dystrophy Association.
ABBREVIATION
- GST
glutathione S-transferase
References
- 1.Rees D J, Ades S E, Singer S J, Hynes R O. Nature (London) 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- 2.Niggli V, Kaufmann S, Goldmann W H, Weber T, Isenberg G. Eur J Biochem. 1994;224:951–957. doi: 10.1111/j.1432-1033.1994.00951.x. [DOI] [PubMed] [Google Scholar]
- 3.Burridge K, Connell L. J Cell Biol. 1983;97:359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 5.Nuckolls G H, Romer L H, Burridge K. J Cell Sci. 1992;102:753–762. doi: 10.1242/jcs.102.4.753. [DOI] [PubMed] [Google Scholar]
- 6.Albiges-Rigo C, Frachet P, Block M. J Cell Sci. 1996;108:3317–3329. doi: 10.1242/jcs.108.10.3317. [DOI] [PubMed] [Google Scholar]
- 7.Sydor A M, Su A L, Wang F -W, Xu A, Jay D G. J Cell Biol. 1996;134:1197–1207. doi: 10.1083/jcb.134.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isenberg G, Goldmann W H. J Muscle Res Cell Motil. 1992;13:587–589. doi: 10.1007/BF01738248. [DOI] [PubMed] [Google Scholar]
- 9.Muguruma M, Matsumura S, Fukazawa T. Biochem Biophys Res Commun. 1990;171:1217–1223. doi: 10.1016/0006-291x(90)90815-5. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann S, Piekenbrock T, Goldmann W H, Baermann M, Isenberg G. FEBS Lett. 1991;284:187–191. doi: 10.1016/0014-5793(91)80681-r. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt J M, Robson R M, Zhang J, Stromer M H. Biochem Biophys Res Commun. 1993;197:660–666. doi: 10.1006/bbrc.1993.2530. [DOI] [PubMed] [Google Scholar]
- 12.Burridge K, Mangeat P. Nature (London) 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz A, Duggan K, Buck C, Beckerle M C, Burridge K. Nature (London) 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- 14.Knezevic I, Leisner I, Lam S C-T. J Biol Chem. 1995;271:16416–16421. doi: 10.1074/jbc.271.27.16416. [DOI] [PubMed] [Google Scholar]
- 15.Chen H C, Appeddu P A, Parsons J T, Hildebrand J D, Schaller M D, Guan J-L. J Biol Chem. 1995;270:16995–16999. doi: 10.1074/jbc.270.28.16995. [DOI] [PubMed] [Google Scholar]
- 16.Goldmann W H, Bremer A, Haner M, Aebi U, Iserberg G. J Struct Biol. 1994;112:3–10. doi: 10.1006/jsbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 17.Muguruma M, Nishimuta S, Tomisaka Y, Tatsumi I, Matsumura S. J Biochem. 1995;117:1036–1042. doi: 10.1093/oxfordjournals.jbchem.a124803. [DOI] [PubMed] [Google Scholar]
- 18.O’Halloran T, Burridge K. Biochim Biophys Acta. 1986;869:337–349. doi: 10.1016/0167-4838(86)90074-9. [DOI] [PubMed] [Google Scholar]
- 19.Hemmings L, Rees D J G, Ohanian V, Bolton S J, Gilmore A P, Patel B, Priddle H, Trevithick J E, Hynes R O, Critchley D R. J Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 20.Belliveau M J, Lutchman M, Claudio J O, Marineau C, Rouleau G A. Biochem Cell Biol. 1995;73:733–737. doi: 10.1139/o95-081. [DOI] [PubMed] [Google Scholar]
- 21.Kreitmeier M, Gerisch G, Heizer C, Mueller-Taubenberger A. J Cell Biol. 1996;129:179–188. doi: 10.1083/jcb.129.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulder G L, Huang M M, Waterston R H, Barstead R J. Mol Biol Cell. 1996;7:1181–1193. doi: 10.1091/mbc.7.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtzman D A, Yang S, Drubin D G. J Cell Biol. 1996;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raths S, Rohrer J, Crausax F, Riezman H. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na S, Hincapie M, McCusker J H, Haber J E. J Biol Chem. 1995;270:6815–6823. doi: 10.1074/jbc.270.12.6815. [DOI] [PubMed] [Google Scholar]
- 26.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Zheng Y, Drubin D G. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spudich J, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 29.MacLean-Fletcher S, Pollard T D. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 30.Kouyama T, Mihashi K. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 31.Cooper J A, Walker S B, Pollard T D. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 32.Cook R K, Blake W T, Rubenstein P A. J Biol Chem. 1992;267:9430–9436. [PubMed] [Google Scholar]
- 33.Molony L, McCaslin D, Abernethy J, Paschal B, Burridge K. J Biol Chem. 1987;262:7790–7795. [PubMed] [Google Scholar]
- 34.Henikoff S, Henikoff J G, Alford W J, Pietrokovski S. Gene-COMBIS. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 35.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 36.Smith H O, Annau T M, Chandrasegaran S. Proc Natl Acad Sci USA. 1990;87:826–830. doi: 10.1073/pnas.87.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff J G. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson R P, Craig S W. Nature (London) 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Robson R M, Schmidt J M, Stromer M H. Biochem Biophys Res Commun. 1996;218:530–537. doi: 10.1006/bbrc.1996.0095. [DOI] [PubMed] [Google Scholar]
- 41.Walker J, Crowley P, Moreman A D, Barrett J. Mol Biochem Parisitol. 1993;61:255–264. doi: 10.1016/0166-6851(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 42.McTigue M A, Williams D R, Tainer J A. J Mol Biol. 1995;246:21–27. doi: 10.1006/jmbi.1994.0061. [DOI] [PubMed] [Google Scholar]
- 43.Turunen O, Wahlstrom T, Vaheri A. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsudaira P. Trends Biochem Sci. 1991;16:87–92. doi: 10.1016/0968-0004(91)90039-x. [DOI] [PubMed] [Google Scholar]
- 45.Bretscher A, Drees B, Harsay E, Schott D, Wang T. J Cell Biol. 1996;126:821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munn A L, Stevenson B J, Geli M I, Riezman H. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]


