Abstract
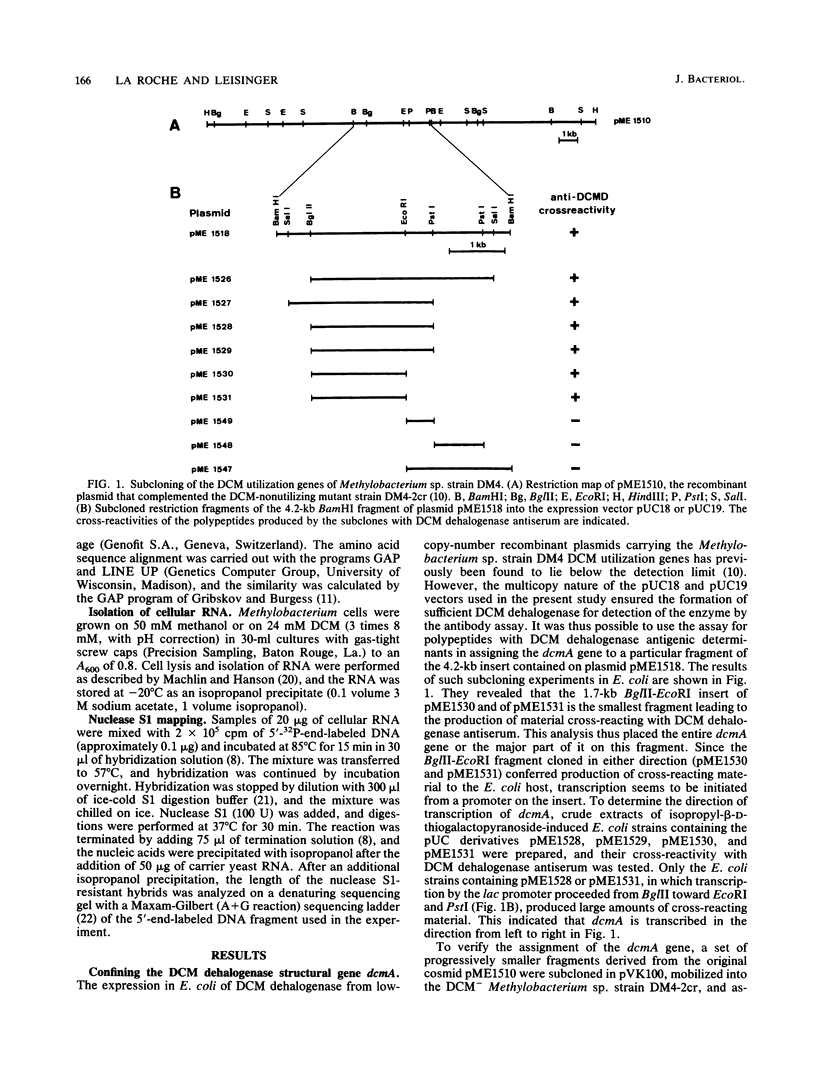
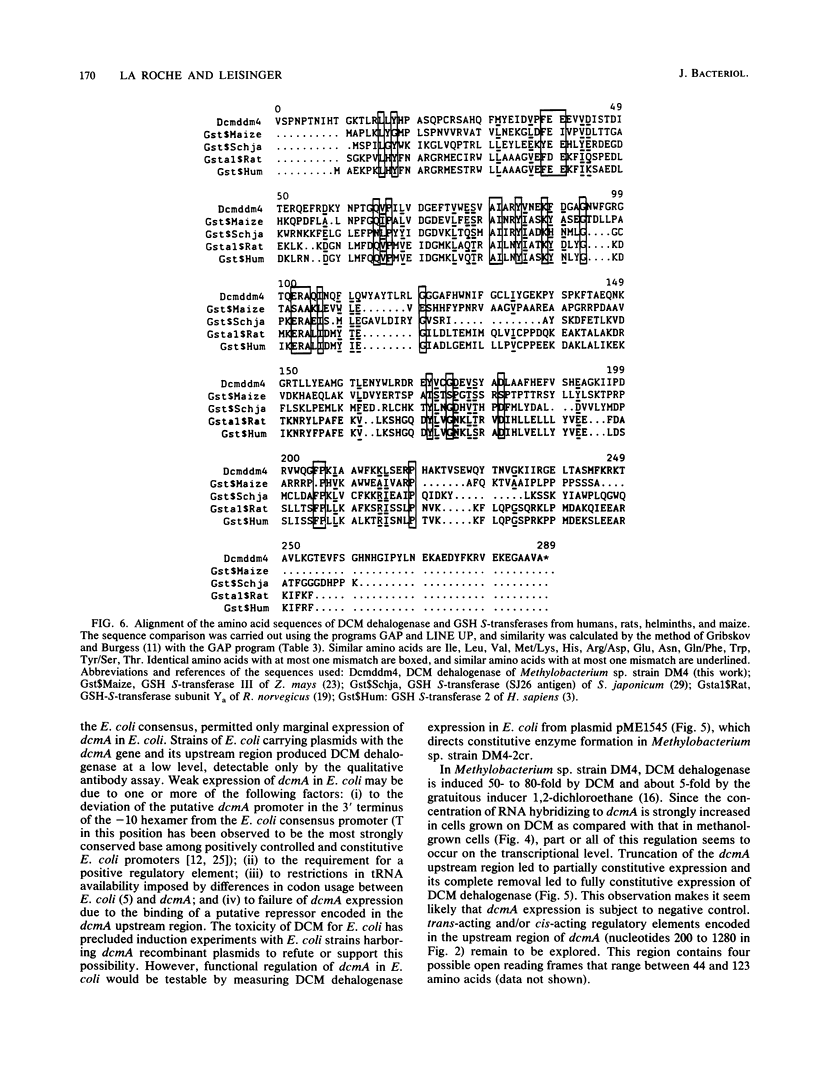
The nucleotide sequence of a cloned 2.8-kilobase-pair BamHI-PstI fragment containing dcmA, the dichloromethane dehalogenase structural gene from Methylobacterium sp. strain DM4, was determined. An open reading frame with a coding capacity of 287 amino acids (molecular weight, 37,430) was identified as dcmA by its agreement with the N-terminal amino acid sequence, the total amino acid composition, and the subunit size of the purified enzyme. Alignment of the deduced dichloromethane dehalogenase amino acid sequence with amino acid sequences of the functionally related eucaryotic glutathione S-transferases revealed three regions containing highly conserved amino acid residues and indicated that dcmA is a member of the glutathione S-transferase supergene family. The 5' terminus of in vivo dcmA transcripts was determined by nuclease S1 mapping to be 82 base pairs upstream of the GTG initiation codon of dcmA. Despite a putative promoter sequence with high resemblance to the Escherichia coli -10 and -35 consensus sequences, located at an appropriate distance from the transcription start point, dcmA was only marginally expressed in E. coli. The strong induction of dichloromethane dehalogenase in Methylobacterium sp. by dichloromethane was abolished by deleting the 1.3-kilobase-pair upstream region of dcmA. Plasmid constructs devoid of this region directed expression of dichloromethane dehalogenase at a constitutively induced level.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. E., Anders M. W. Metabolism of dihalomethanes to formaldehyde and inorganic halide--II. Studies on the mechanism of the reaction. Biochem Pharmacol. 1978;27(16):2021–2025. doi: 10.1016/0006-2952(78)90061-8. [DOI] [PubMed] [Google Scholar]
- Arca P., Rico M., Braña A. F., Villar C. J., Hardisson C., Suárez J. E. Formation of an adduct between fosfomycin and glutathione: a new mechanism of antibiotic resistance in bacteria. Antimicrob Agents Chemother. 1988 Oct;32(10):1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board P. G., Webb G. C. Isolation of a cDNA clone and localization of human glutathione S-transferase 2 genes to chromosome band 6p12. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2377–2381. doi: 10.1073/pnas.84.8.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ilio C., Aceto A., Piccolomini R., Allocati N., Faraone A., Cellini L., Ravagnan G., Federici G. Purification and characterization of three forms of glutathione transferase from Proteus mirabilis. Biochem J. 1988 Nov 1;255(3):971–975. doi: 10.1042/bj2550971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälli R., Leisinger T. Plasmid analysis and cloning of the dichloromethane-utilization genes of Methylobacterium sp. DM4. J Gen Microbiol. 1988 Apr;134(4):943–952. doi: 10.1099/00221287-134-4-943. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke H., Günther I., Binder F. A novel cloning vector for the direct selection of recombinant DNA in E. coli. Gene. 1982 Sep;19(2):231–234. doi: 10.1016/0378-1119(82)90011-7. [DOI] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kohler-Staub D., Leisinger T. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J Bacteriol. 1985 May;162(2):676–681. doi: 10.1128/jb.162.2.676-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Grove G., Tu C. P. Cloning and sequence analysis of a cDNA for a rat liver glutathione S-transferase Yb subunit. Nucleic Acids Res. 1986 Aug 11;14(15):6101–6114. doi: 10.1093/nar/14.15.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Li N., Weiss M. J., Reddy C. C., Tu C. P. The nucleotide sequence of a rat liver glutathione S-transferase subunit cDNA clone. J Biol Chem. 1984 May 10;259(9):5536–5542. [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore R. E., Davies M. S., O'Connell K. M., Harding E. I., Wiegand R. C., Tiemeier D. C. Cloning and expression of a cDNA encoding a maize glutathione-S-transferase in E. coli. Nucleic Acids Res. 1986 Sep 25;14(18):7227–7235. doi: 10.1093/nar/14.18.7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz R., Wackett L. P., Egli C., Cook A. M., Leisinger T. Dichloromethane dehalogenase with improved catalytic activity isolated from a fast-growing dichloromethane-utilizing bacterium. J Bacteriol. 1988 Dec;170(12):5698–5704. doi: 10.1128/jb.170.12.5698-5704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Davern K. M., Board P. G., Tiu W. U., Garcia E. G., Mitchell G. F. Mr 26,000 antigen of Schistosoma japonicum recognized by resistant WEHI 129/J mice is a parasite glutathione S-transferase. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8703–8707. doi: 10.1073/pnas.83.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Holloway B. W. A mutant sex factor of Pseudomonas aeruginosa. Genet Res. 1972 Feb;19(1):91–108. doi: 10.1017/s0016672300014294. [DOI] [PubMed] [Google Scholar]
- Suguoka Y., Kano T., Okuda A., Sakai M., Kitagawa T., Muramatsu M. Cloning and the nucleotide sequence of rat glutathione S-transferase P cDNA. Nucleic Acids Res. 1985 Sep 11;13(17):6049–6057. doi: 10.1093/nar/13.17.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., Rodkey J. A., Bennett C. D., Lu A. Y., Pickett C. B. Rat liver glutathione S-transferases. Construction of a cDNA clone complementary to a Yc mRNA and prediction of the complete amino acid sequence of a Yc subunit. J Biol Chem. 1985 May 10;260(9):5820–5825. [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., Rothkopf G. S., Pickett C. B. Structural analysis of a rat liver glutathione S-transferase Ya gene. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9393–9397. doi: 10.1073/pnas.83.24.9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Qian B. Human liver glutathione S-transferases: complete primary sequence of an Ha subunit cDNA. Biochem Biophys Res Commun. 1986 Nov 26;141(1):229–237. doi: 10.1016/s0006-291x(86)80358-8. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]