Abstract
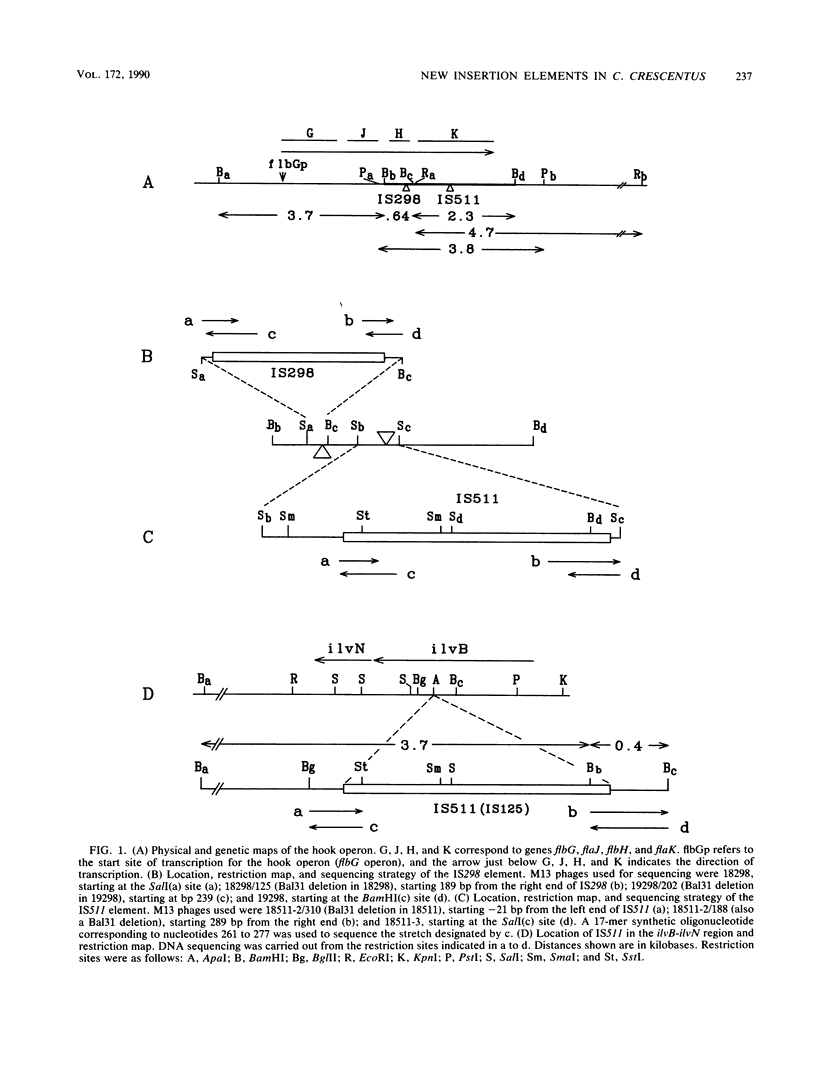
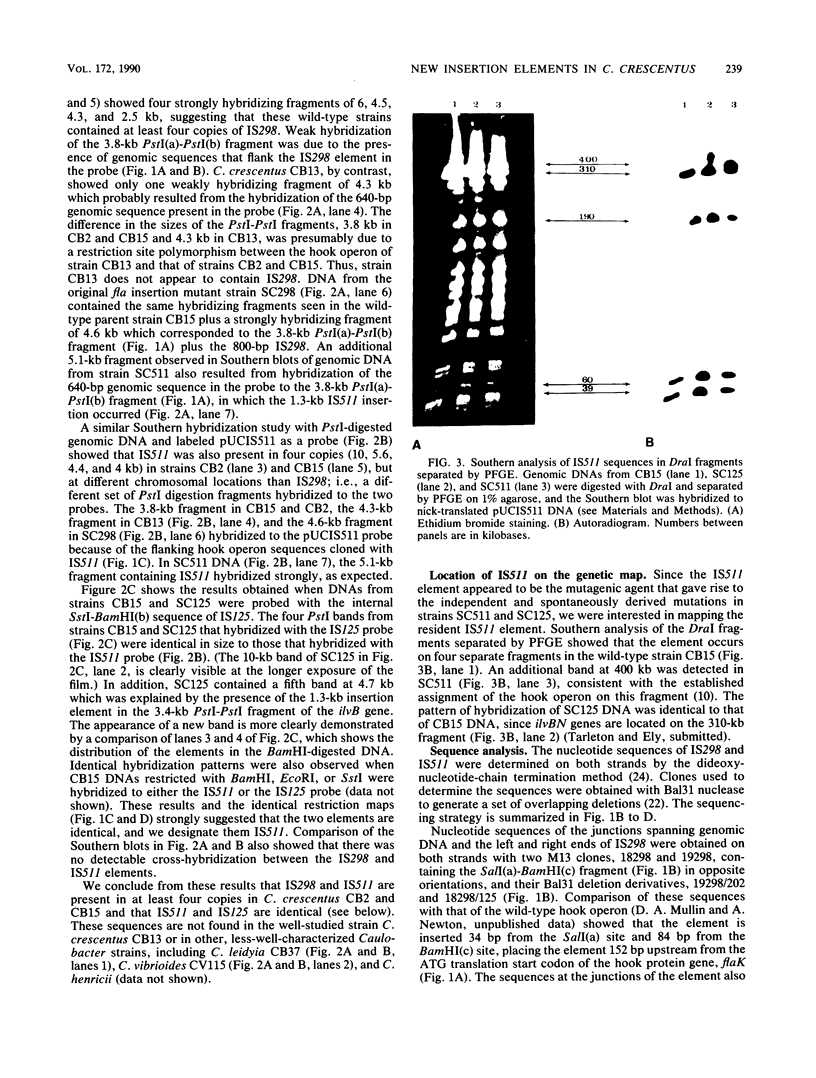
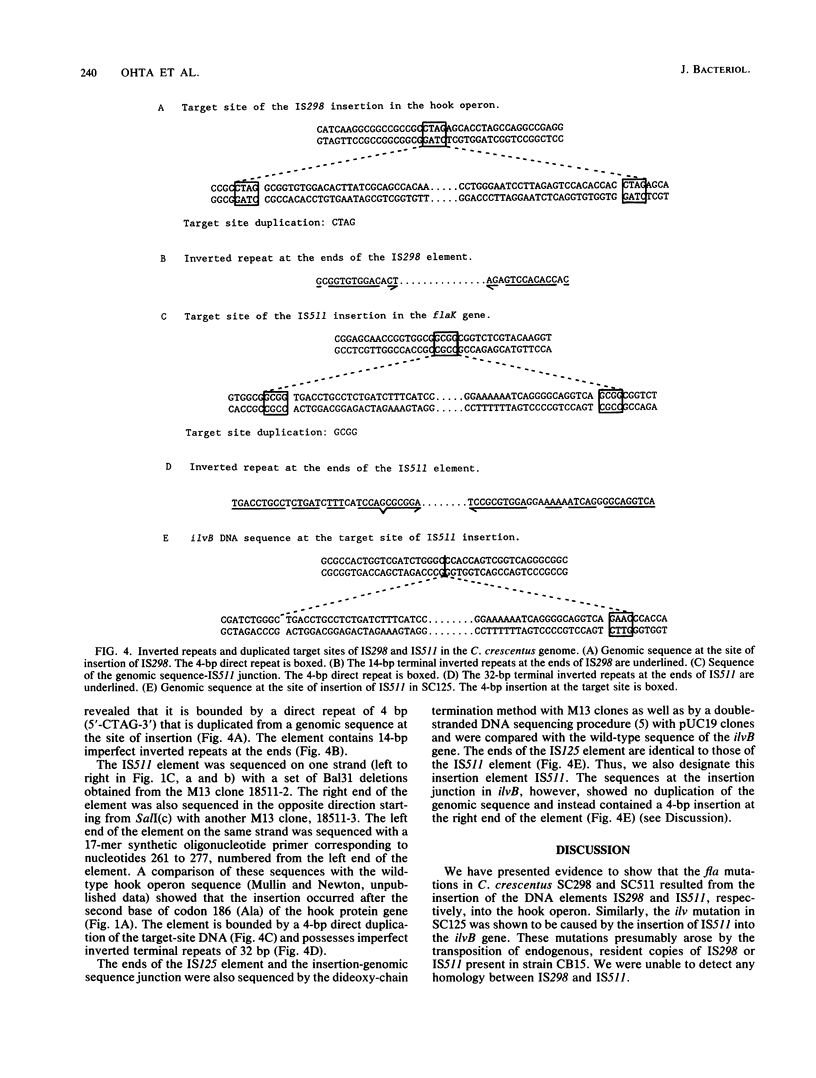
We describe two insertion elements isolated from Caulobacter crescentus that are designated IS298 and IS511. These insertion elements were cloned from spontaneous flagellar (fla) gene mutants SC298 and SC511 derived from the wild-type strain CB15 (ATCC 19089), in which they were originally identified as insertions in the flbG operon of the hook gene cluster (N. Ohta, E. Swanson, B. Ely, and A. Newton, J. Bacteriol. 158:897-904, 1984). IS298 and IS511 were each present in C. crescentus CB2 and CB15 in at least four different positions, but neither was present in strain CB13 or in several Caulobacter species examined, including C. vibrioides, C. leidyia, and C. henricii. Nucleotide sequence analysis across the chromosome-insertion element junctions showed that IS298 is located 152 base pairs (bp) upstream from the ATG translation start of the hook protein gene flaK, where it is bounded by a 4-bp direct repeat derived from the site of insertion, and that IS511 is inserted at codon 186 of the flaK coding sequence, where it is also bounded by a 4-bp direct repeat duplicated from the site of insertion. The ilvB102 mutation in strain SC125 was also shown to result from insertion sequence IS511, but no duplication of the genomic sequence was present at the insertion element junctions. IS298 contains an imperfect terminal inverted repeat 16 bp long, and IS511 contains a 32-bp inverted repeat at the termini. IS298 and IS511 are the first insertion elements described in C. crescentus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auerswald E. A., Ludwig G., Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Aroca E., Mendiola M. V., Zabala J. C., de la Cruz F. Transposition of IS91 does not generate a target duplication. J Bacteriol. 1987 Jan;169(1):442–443. doi: 10.1128/jb.169.1.442-443.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988 Sep 7;68(2):323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Murphy E., Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984 Jan 19;307(5948):292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- Ohta N., Swanson E., Ely B., Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984 Jun;158(3):897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]