Abstract
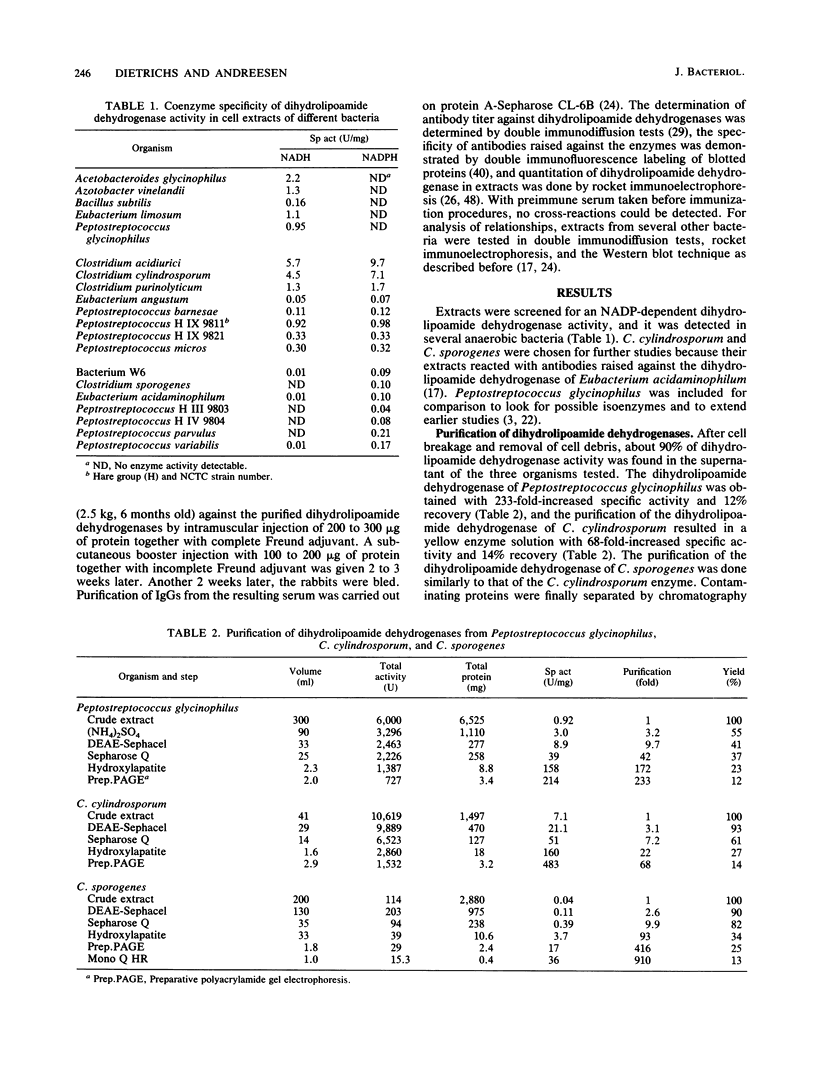
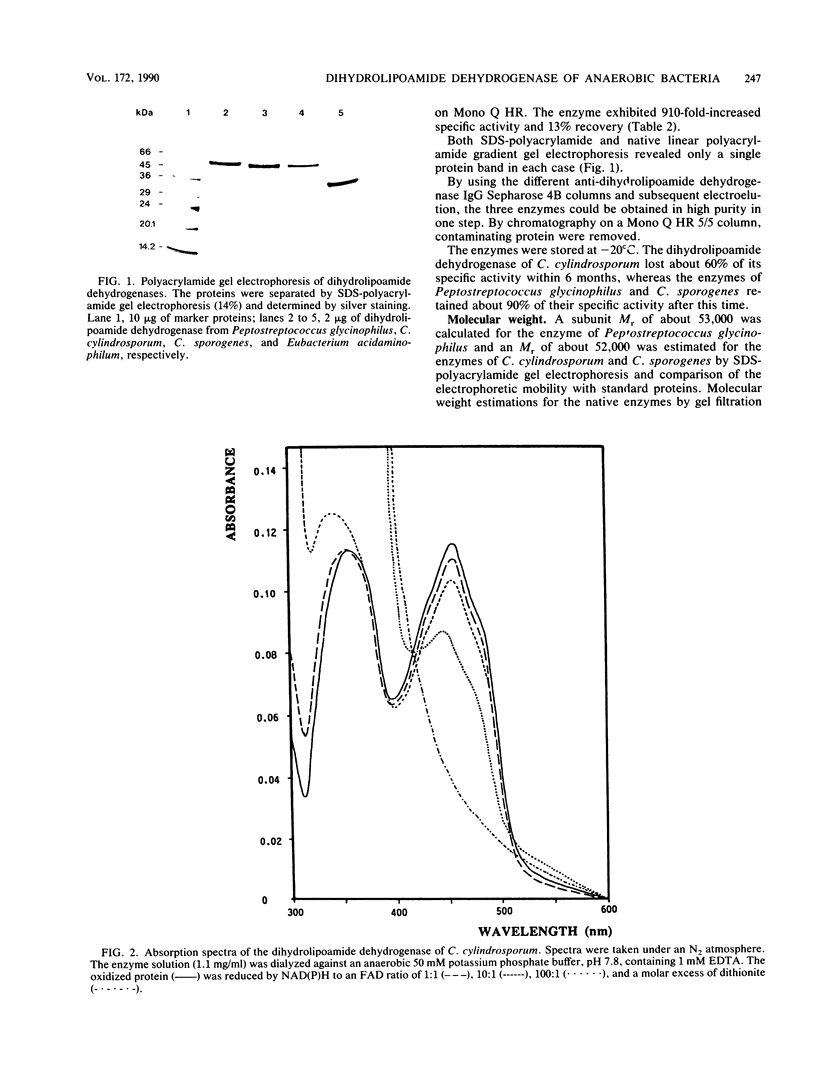
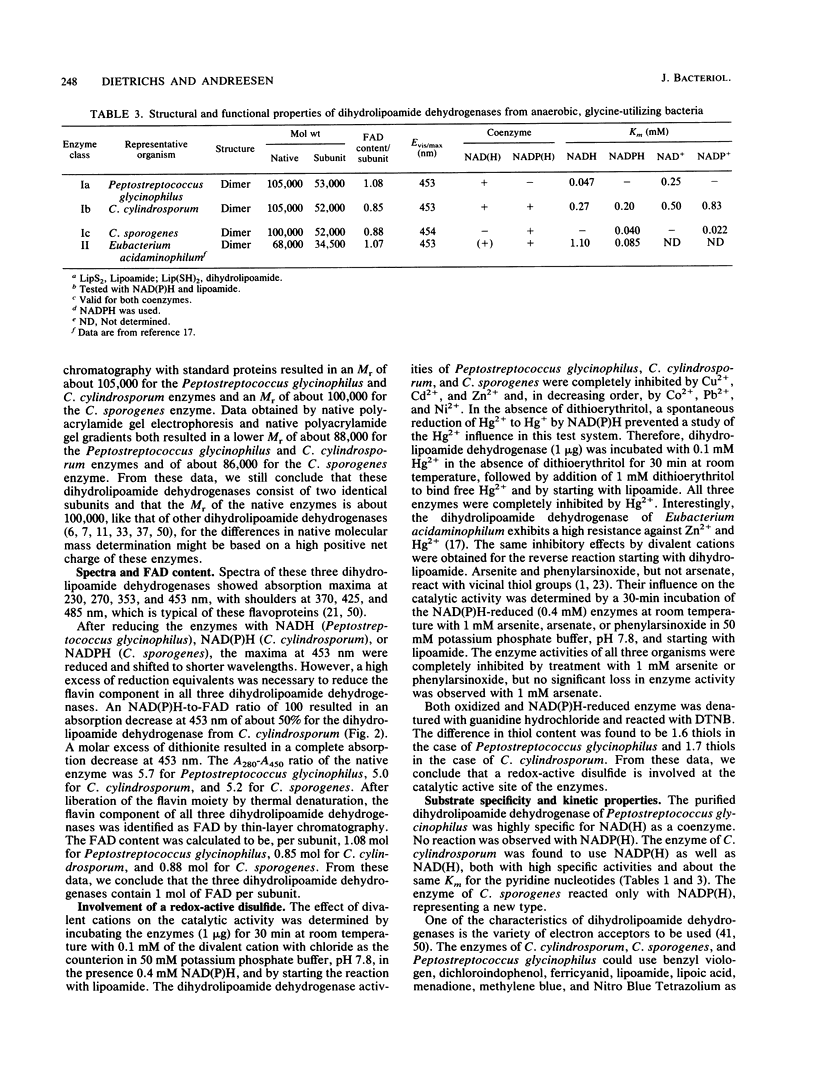
Three different dihydrolipoamide dehydrogenases were purified to homogenity from the anaerobic glycine-utilizing bacteria Clostridium cylindrosporum, Clostridium sporogenes, and Peptostreptococcus glycinophilus, and their basic properties were determined. The enzyme isolated from P. glycinophilus showed the properties typical of dihydrolipoamide dehydrogenases: it was a dimer with a subunit molecular mass of 53,000 and contained 1 mol of flavin adenine dinucleotide and 2 redox-active sulfhydryl groups per subunit. Only NADH was active as a coenzyme for reduction of lipoamide. Spectra of the oxidized enzyme exhibited maxima at 230, 270, 353, and 453 nm, with shoulders at 370, 425, and 485 nm. The dihydrolipoamide dehydrogenases of C. cylindrosporum and C. sporogenes were very similar in their structural properties to the enzyme of P. glycinophilus except for their coenzyme specificity. The enzyme of C. cylindrosporum used NAD(H) as well as NADP(H), whereas the enzyme of C. sporogenes reacted only with NADP(H), and no reaction could be detected with NAD(H). Antibodies raised against the dihydrolipoamide dehydrogenase of C. cylindrosporum reacted with extracts of Clostridium acidiurici, Clostridium purinolyticum, and Eubacterium angustum, whereas antibodies raised against the enzymes of P. glycinophilus and C. sporogenes showed no cross-reaction with extracts from 42 organisms tested.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamson S. R., Robinson J. A., Stevenson K. J. Inhibition of pyruvate dehydrogenase multienzyme complex from Escherichia coli with a radiolabeled bifunctional arsenoxide: evidence for an essential histidine residue at the active site of lipoamide dehydrogenase. Biochemistry. 1984 Mar 13;23(6):1269–1274. doi: 10.1021/bi00301a039. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Sokatch J. R. Sequence analysis of the lpdV gene for lipoamide dehydrogenase of branched-chain-oxoacid dehydrogenase of Pseudomonas putida. Eur J Biochem. 1989 Jan 15;179(1):61–69. doi: 10.1111/j.1432-1033.1989.tb14521.x. [DOI] [PubMed] [Google Scholar]
- Burns G., Sykes P. J., Hatter K., Sokatch J. R. Isolation of a third lipoamide dehydrogenase from Pseudomonas putida. J Bacteriol. 1989 Feb;171(2):665–668. doi: 10.1128/jb.171.2.665-668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers D. J., Pons G., Patel M. S. Dihydrolipoamide dehydrogenase: functional similarities and divergent evolution of the pyridine nucleotide-disulfide oxidoreductases. Arch Biochem Biophys. 1989 Feb 1;268(2):409–425. doi: 10.1016/0003-9861(89)90309-3. [DOI] [PubMed] [Google Scholar]
- Costilow R. N. Selenium requirement for the growth of Clostridium sporogenes with glycine as the oxidant in stickland reaction systems. J Bacteriol. 1977 Jul;131(1):366–368. doi: 10.1128/jb.131.1.366-368.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J. Archaebacteria: the comparative enzymology of their central metabolic pathways. Adv Microb Physiol. 1988;29:165–231. doi: 10.1016/s0065-2911(08)60348-3. [DOI] [PubMed] [Google Scholar]
- Danson M. J., Eisenthal R., Hall S., Kessell S. R., Williams D. L. Dihydrolipoamide dehydrogenase from halophilic archaebacteria. Biochem J. 1984 Mar 15;218(3):811–818. doi: 10.1042/bj2180811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürre P., Andreesen J. R. Purine and glycine metabolism by purinolytic clostridia. J Bacteriol. 1983 Apr;154(1):192–199. doi: 10.1128/jb.154.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürre P., Andreesen J. R. Selenium-dependent growth and glycine fermentation by Clostridium purinolyticum. J Gen Microbiol. 1982 Jul;128(7):1457–1466. doi: 10.1099/00221287-128-7-1457. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Freudenberg W., Andreesen J. R. Purification and partial characterization of the glycine decarboxylase multienzyme complex from Eubacterium acidaminophilum. J Bacteriol. 1989 Apr;171(4):2209–2215. doi: 10.1128/jb.171.4.2209-2215.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg W., Dietrichs D., Lebertz H., Andreesen J. R. Isolation of an atypically small lipoamide dehydrogenase involved in the glycine decarboxylase complex from Eubacterium acidaminophilum. J Bacteriol. 1989 Mar;171(3):1346–1354. doi: 10.1128/jb.171.3.1346-1354.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason F. K., Holmgren A. Thioredoxin and related proteins in procaryotes. FEMS Microbiol Rev. 1988 Dec;4(4):271–297. doi: 10.1111/j.1574-6968.1988.tb02747.x. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Holmgren A. Pyridine nucleotide - disulfide oxidoreductases. Experientia Suppl. 1980;36:149–180. doi: 10.1007/978-3-0348-5419-1_5. [DOI] [PubMed] [Google Scholar]
- Klein S. M., Sagers R. D. Glycine metabolism. 3. A flavin-linked dehydrogenase associated with the glycine cleavage system in Peptococcus glycinophilus. J Biol Chem. 1967 Jan 25;242(2):297–300. [PubMed] [Google Scholar]
- Knowles F. C. Reactions of lipoamide dehydrogenase and glutathione reductase with arsonic acids and arsonous acids. Arch Biochem Biophys. 1985 Oct;242(1):1–10. doi: 10.1016/0003-9861(85)90472-2. [DOI] [PubMed] [Google Scholar]
- Kohring G. W., Mayer F., Mayer H. Immunoelectron microscopic localization of the restriction endonuclease EcoRI in Escherichia coli BS 5. Eur J Cell Biol. 1985 May;37:1–6. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laurell C. B. Quantitative estimation of proteins by electrophoresis in agarose gel containing antibodies. Anal Biochem. 1966 Apr;15(1):45–52. doi: 10.1016/0003-2697(66)90246-6. [DOI] [PubMed] [Google Scholar]
- Margolis J., Kenrick K. C. Polyacrylamide gel-electrophoresis across a molecular sieve gradient. Nature. 1967 Jun 24;214(5095):1334–1336. doi: 10.1038/2141334a0. [DOI] [PubMed] [Google Scholar]
- McCully V., Burns G., Sokatch J. R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa PAO. Biochem J. 1986 Feb 1;233(3):737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigiet V. P., Conley R. R. Purification of thioredoxin, thioredoxin reductase, and glutathione reductase by affinity chromatography. J Biol Chem. 1977 Sep 25;252(18):6367–6372. [PubMed] [Google Scholar]
- SAGERS R. D., GUNSALUS I. C. Intermediatry metabolism of Diplococcus glycinophilus. I. Glycine cleavage and one-carbon interconversions. J Bacteriol. 1961 Apr;81:541–549. doi: 10.1128/jb.81.4.541-549.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefer-Ullrich H., Wagner R., Dürre P., Andreesen J. R. Comparative studies on physiology and taxonomy of obligately purinolytic clostridia. Arch Microbiol. 1984 Aug;138(4):345–353. doi: 10.1007/BF00410902. [DOI] [PubMed] [Google Scholar]
- Scouten W. H., McManus I. R. Microbial lipoamide dehydrogenase. Purification and some characteristics of the enzyme derived from selected microorganisms. Biochim Biophys Acta. 1971 Feb 10;227(2):248–263. doi: 10.1016/0005-2744(71)90058-1. [DOI] [PubMed] [Google Scholar]
- Shames S. L., Fairlamb A. H., Cerami A., Walsh C. T. Purification and characterization of trypanothione reductase from Crithidia fasciculata, a newly discovered member of the family of disulfide-containing flavoprotein reductases. Biochemistry. 1986 Jun 17;25(12):3519–3526. doi: 10.1021/bi00360a007. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., Burns G. Oxidation of glycine by Pseudomonas putida requires a specific lipoamide dehydrogenase. Arch Biochem Biophys. 1984 Feb 1;228(2):660–666. doi: 10.1016/0003-9861(84)90036-5. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Gebrosky J., Sokatch D. J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Sundquist A. R., Fahey R. C. The novel disulfide reductase bis-gamma-glutamylcystine reductase and dihydrolipoamide dehydrogenase from Halobacterium halobium: purification by immobilized-metal-ion affinity chromatography and properties of the enzymes. J Bacteriol. 1988 Aug;170(8):3459–3467. doi: 10.1128/jb.170.8.3459-3467.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow R. D., Setlow P. Purification and characterization of a Bacillus megaterium disulfide reductase specific for disulfides containing pantethine 4',4"-diphosphate. J Bacteriol. 1983 Jan;153(1):475–484. doi: 10.1128/jb.153.1.475-484.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. S., Wand A. J., Templeton D. M., Weiss P. M. Multifunctionality of lipoamide dehydrogenase promotion of electron transferase reaction. Arch Biochem Biophys. 1983 Sep;225(2):554–561. doi: 10.1016/0003-9861(83)90067-x. [DOI] [PubMed] [Google Scholar]
- Turner D. C., Stadtman T. C. Purification of protein components of the clostridial glycine reductase system and characterization of protein A as a selenoprotein. Arch Biochem Biophys. 1973 Jan;154(1):366–381. doi: 10.1016/0003-9861(73)90069-6. [DOI] [PubMed] [Google Scholar]
- Uyeda K., Rabinowitz J. C. Enzymes of clostridial purine fermentation. Methylenetetrahydrofolate dehydrogenase. J Biol Chem. 1967 Oct 10;242(19):4378–4385. [PubMed] [Google Scholar]
- VALENTINE R. C., BRILL W. J., SAGERS R. D. FERREDOXIN LINKED DPN REDUCTION BY PYRUVATE IN EXTRACTS OF CLOSTRIDIUM ACIDI-URICI. Biochem Biophys Res Commun. 1963 Aug 1;12:315–319. doi: 10.1016/0006-291x(63)90303-6. [DOI] [PubMed] [Google Scholar]
- Wagner R., Andreesen J. R. Differentiation between Clostridium acidiurici and Clostridium cylindrosporum on the basis of specific metal requirements for formate dehydrogenase formation. Arch Microbiol. 1977 Sep 28;114(3):219–224. doi: 10.1007/BF00446865. [DOI] [PubMed] [Google Scholar]
- Walker J. L., Oliver D. J. Glycine decarboxylase multienzyme complex. Purification and partial characterization from pea leaf mitochondria. J Biol Chem. 1986 Feb 15;261(5):2214–2221. [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weeke B. Rocket immunoelectrophoresis. Scand J Immunol Suppl. 1973;1:37–46. doi: 10.1111/j.1365-3083.1973.tb03777.x. [DOI] [PubMed] [Google Scholar]
- Westphal A. H., de Kok A. Lipoamide dehydrogenase from Azotobacter vinelandii. Molecular cloning, organization and sequence analysis of the gene. Eur J Biochem. 1988 Mar 1;172(2):299–305. doi: 10.1111/j.1432-1033.1988.tb13887.x. [DOI] [PubMed] [Google Scholar]