Abstract
Self-recognition, self-selection, and dynamic self-organization are of fundamental importance for the assembly of all supramolecular systems, but molecular-level information is not generally accessible. We present direct examples of these critical steps by using scanning tunneling microscopy to study mixtures of complementary organic ligands on a copper substrate. The ligands coordinate cooperatively with iron atoms to form well ordered arrays of rectangular multicomponent compartments whose size and shape can be deliberately tuned by selecting ligands of desired length from complementary ligand families. We demonstrate explicitly that highly ordered supramolecular arrays can be produced from redundant ligand mixtures by molecular self-recognition and -selection, enabled by efficient error correction and cooperativity, and show an example of failed self-selection due to error tolerance in the ligand mixture, leading to a disordered structure.
Keywords: nanostructure, scanning tunneling microscopy, self-assembly, surface chemistry, organic molecule ligands
Supramolecular self-organization, directed by information stored in molecular components and read out through their specific interactions, represents the pivotal operation in the spontaneous but controlled build-up of structurally organized and functionally integrated molecular systems (1). Metal–ligand coordination bonding is an effective strategy for strong, directional bonding to stabilize designed, self-assembled supramolecular architectures (2–5). Substrate-supported, 2D supramolecular coordination for efficient nanometer-scale patterning of solid surfaces has been demonstrated (6, 7). By selecting organic ligands with appropriate size, geometry, and binding moieties, specific tailored architectures can be produced across a substrate completely by self-assembly of the molecules. Such patterning of surfaces is of great interest for potential applications in surface nanofunctionalization, templated growth, and controlling 2D molecular nanoarrays. Molecular level insight has provided structural details of these systems, and here we explore multicomponent systems at that level to illustrate critical assembly requirements for these systems and (bio-)molecular systems in general.
Self-selection occurs when the involved molecular components are sufficiently instructed to allow self-recognition and -assembly into discrete supramolecular architectures (8). Processes of modular self-assembly, dynamic self-organization, and self-selection are of fundamental importance for the assembly of all supramolecular systems, but molecular-level information is not generally accessible. Using scanning tunneling microscopy (STM), we address these issues by studying mixtures of complementary organic molecule ligands on a copper substrate that coordinate cooperatively with iron atoms to form regular arrays of rectangular multicomponent compartments. Ensembles of these complementary components serve as model systems to investigate the dynamic bottom-up self-organization process of modular multicomponent systems with nanometer accuracy. These experiments provide evidence for cooperativity, self-recognition, and self-selection.
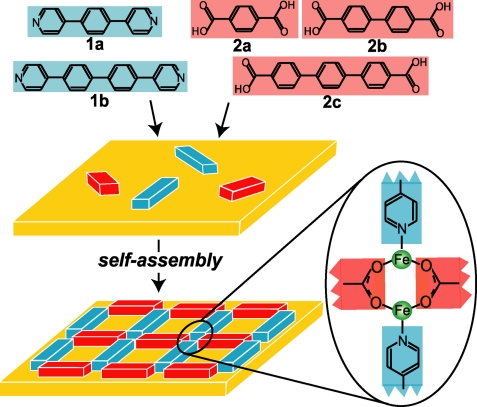
Surface-supported open-network structures can be generated from molecular components by (i) hydrogen bond directed homo- (9, 10) and heteroassembly (11) of organic molecules or by (ii) the coordination of metal ions with organic molecules acting as ligands (12, 13). For example, linear polyaromatic bis-carboxylic acids (e.g., ligands 2a, 2b, and 2c in Fig. 1) form, in combination with Fe centers on Cu(100) surfaces, extended domains of homoligand coordination networks (7, 14). It was shown that a ligand containing a carboxylic group at one end and a pyridine group at the opposite end coordinates to a Fe dimer through bridging μ2-carboxylates and axial pyridyls, leading to coordination centers of [(Fe2)(carboxylate)2/2(pyridyl)2/2]n stoichiometry, as illustrated in the magnified illustration provided in Fig. 1 (15).
Fig. 1.
Complementary molecular ligands deposited with Fe atoms on Cu(100) self-assemble into regular rectangular arrays. Schematic illustration of molecules used in this study (top). Molecules and Fe atoms deposited to surface and annealed at 450 K to activate mobility for assembly. Also shown is a detail of local bonding arrangement at each node in the rectangular networks.
In the following, we study the coordination of iron atoms by the members of two series of organic ligands, linear bis-carboxylic acids and bipyridines, under 2D confinement, i.e., on a Cu(100) surface in ultrahigh vacuum conditions (Fig. 1). Two bipyridines (ligand 1a, 1,4-bipyridyl-benzene; ligand 1b, 4,4′-bipyridyl-biphenyl) and three bis-carboxylic acids (ligand 2a, 1,4-benzoic-dicarboxylic acid; ligand 2b, 1,4′-biphenyl-dicarboxylic acid; ligand 2c, 4,1′,4′,1′-terphenyl-1,4′-dicarboxylic acid) were used during the surface-confined metal coordination reactions (Fig. 1). All involved organic ligands are of polyaromatic nature and can be considered as rigid rods, each of a unique length, enabling linear, terminal coordination of metal centers at discrete distances in the range of 0.8 nm to 2.6 nm.
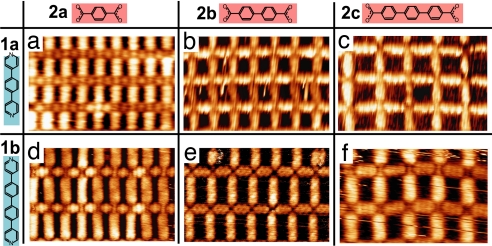
In a first experiment, we studied the self-assembly of each of the six possible binary heteroligand combinations (i.e., one ligand each from families 1 and 2). (We use the phrase “binary mixture” to refer to a mixture of two ligand species with Fe atoms, not one ligand plus Fe.) The codeposition of any of the binary combinations with iron atoms can yield highly ordered, extended coordination networks (Fig. 2) with the appropriate absolute quantities and ratios of deposited components. The internal dimensions of the rectangular compartments can be varied in a modular way through the backbone length of the selected molecular ligands leading to open cavity areas ranging from 1.9 nm2 to 4.2 nm2 (Fig. 2).
Fig. 2.
Steering the size and aspect ratio of rectangular molecular-scale compartments via the backbone length of the ligands in self-assembled iron coordination networks. STM images show six possible binary combinations [(Fe2)(1)2/2(2)2/2]n of bipyridine (ligands 1a and 1b) and bis-carboxylic acid (ligands 2a, 2b, and 2c) ligands. All images are 9.4 × 6.0 nm. Structure periodicity is 1.1 × 1.8 nm (a), 1.5 × 1.8 nm (b), 1.8 × 1.8 nm (c), 1.1 × 2.3 nm (d), 1.5 × 2.3 nm (e), and 1.8 × 2.3 nm (f).
The stoichiometry at each node of these binary coordination networks is invariably represented by [(Fe2)(1)2/2(2)2/2] (magnification in Fig. 1). In terms of self-organization, three unique components (Fe, 1, 2) are associated to each six-membered heteroassembly coordination node. Despite the possibility to form homotopic carboxylate-Fe coordination, the preference of the heterotopic coordination mode over homotopic or alternative structures demonstrates the robustness of the [(Fe2)(1)2/2(2)2/2]n coordination motif. The ligands follow the principle of “maximal site occupancy” by realizing metal coordination at all available sites (1, 16); however, this is not fully achieved by the iron centers, because apical coordination is obviously impossible due to the specific steric demands at the surface (17, 18).
The robust self-assembly of the 2D supramolecular Fe/1/2 system sets the stage for the investigation of dynamic aspects of the system. The formation of “instructed” mixtures allows multiple and parallel processing (e.g., self-recognition and self-selection) of the structural and electronic information stored in the ensemble of components (ligands, metal centers, coordination algorithms, etc.) (1, 8, 16). Additionally, the presence of a substrate imposes well-defined singularities: (i) The freedom of molecular movement is strictly confined to 2D, (ii) molecular conformational freedom and supramolecular coordination algorithms are changed in comparison to bulk materials, and (iii) the interaction of the evolving supramolecular structure with the underlying metallic surface imposes electronic screening (19).
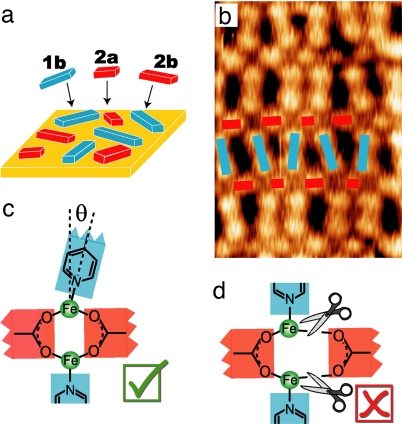
The ternary combination of one bipyridine ligand 1b and two bis-carboxylic acid ligands 2a and 2b was codeposited with iron centers on the Cu(100) surface (≈4:2:1:1 number ratio of Fe/1b/2a/2b). The mixture leads to small domains of distorted network arrangement, indicating a low level of structural accomplishment, even after thermal annealing at 450 K (Fig. 3b). A closer inspection of the STM data reveals that the bis-carboxylates 2a and 2b are strictly linked within horizontal rows (red bars), whereas the columns are exclusively formed by the bipyridine ligand 1b (blue bars). The linear sequence of the bis-carboxylates 2a and 2b is randomly distributed within each row and uncorrelated to the succession of molecules in the neighboring rows. Structural adaptation to this random sequence is accomplished by the bipyridine ligands 1b through significant deviations in the pyridine–Fe-dimer angle, as shown in Fig. 3c (θ falls in a range of ±15°). Obviously, the multicomponent mixture Fe/1b/2a/2b is hindered from reaching the thermodynamically favored structural phase, i.e., regular arrays of rectangular compartments Fe/1b/2a and Fe/1b/2b.
Fig. 3.
Distorted rectangular coordination network exhibiting structural error tolerance. (a) The ternary ligand combination 1b/2a/2b was codeposited with Fe on a Cu(100) surface. (b) STM image showing the structural disorder within the 2D coordination network. Image size is 6.9 × 9.6 nm. (c) Schematic representation of the local coordination structure at the nodes of the network, illustrating a distortion of the pyridyl–Fe bond angle to accommodate the random packing of ligands 2a and 2b. (c and d) This bond distortion (c) is favored compared with breaking the double C–O Fe bond (d), which would be necessary for self-selection and ordering of ligands 2a and 2b (blue, bipyridine ligand 1b; red, bis-carboxylates 2a or 2b; dashed lines to indicate tilting of ligand 1b by angle θ from perpendicular structure).
Fe bond (d), which would be necessary for self-selection and ordering of ligands 2a and 2b (blue, bipyridine ligand 1b; red, bis-carboxylates 2a or 2b; dashed lines to indicate tilting of ligand 1b by angle θ from perpendicular structure).
The metastable structure can be understood by considering the local adaptivity of the coordination schemes. Among the ligands coordinated to the Fe dimer node (Fig. 3c), the negatively charged bridging η2 bis-carboxylates can be considered as the less labile ligands in comparison with the (only) monodentately bound, neutral bipyridyl ligands. Although small rotation of the bridging η2 bis-carboxylate–Fe dimer coordination would have a significant energy cost due to the weakening or even loss of one of the C–O Fe coordination bonds [each ≈1.2 eV in vacuum (S. Fabris, personal communication)‖] (see Fig. 4d), the monodentate bipyridyl–Fe coordination [≈1.3 eV in vacuum (S. Fabris, personal communication)¶] is relatively flexible in that such a significant energy cost for changing its angle (as in Fig. 3c) is not expected (probably a small fraction of the bond energy). However, sufficient binding lability is necessary in both cases to allow reversibility of the coordination bond formation, a basic requirement for self-recognition directed self-assembly, giving the systems the ability to undergo error correction and self-repair. As the sample cools down from the 450 K annealing temperature to room temperature, random distributions of ligands 2a and 2b are frozen in the bis-carboxylate–Fe rows when the reversibility of the carboxylate–Fe bonds is suppressed, whereas the more labile monodentate pyridine–Fe bond adapts to (i.e., energetically accommodates) the given sequence of 2a/2b by tilting the bipyridine–Fe-dimer angle θ, leading to distorted rectangular compartments. Bending of the pyridyl–Fe bond (Fig. 3c), allowing an irregular structure, is favorable to rearrangement of the carboxylate ligands, which would require breaking the double C–O
Fe coordination bonds [each ≈1.2 eV in vacuum (S. Fabris, personal communication)‖] (see Fig. 4d), the monodentate bipyridyl–Fe coordination [≈1.3 eV in vacuum (S. Fabris, personal communication)¶] is relatively flexible in that such a significant energy cost for changing its angle (as in Fig. 3c) is not expected (probably a small fraction of the bond energy). However, sufficient binding lability is necessary in both cases to allow reversibility of the coordination bond formation, a basic requirement for self-recognition directed self-assembly, giving the systems the ability to undergo error correction and self-repair. As the sample cools down from the 450 K annealing temperature to room temperature, random distributions of ligands 2a and 2b are frozen in the bis-carboxylate–Fe rows when the reversibility of the carboxylate–Fe bonds is suppressed, whereas the more labile monodentate pyridine–Fe bond adapts to (i.e., energetically accommodates) the given sequence of 2a/2b by tilting the bipyridine–Fe-dimer angle θ, leading to distorted rectangular compartments. Bending of the pyridyl–Fe bond (Fig. 3c), allowing an irregular structure, is favorable to rearrangement of the carboxylate ligands, which would require breaking the double C–O Fe coordination bonds (Fig. 3d). Because those already formed bis-carboxylate–Fe rows of 2a/2b cannot be corrected by the pyridine–Fe bonds, the Fe/1b/2a/2b mixture does not achieve the necessary self-selection of ligands 2a and 2b.
Fe coordination bonds (Fig. 3d). Because those already formed bis-carboxylate–Fe rows of 2a/2b cannot be corrected by the pyridine–Fe bonds, the Fe/1b/2a/2b mixture does not achieve the necessary self-selection of ligands 2a and 2b.
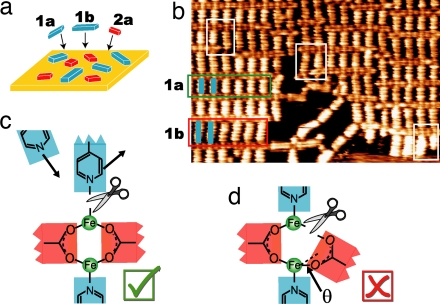
Fig. 4.
Highly ordered coordination networks generated by efficient ligand self-selection. (a) The ternary ligand combination 1a/1b/2a was codeposited with Fe on a Cu(100) surface. (b) STM image showing the local segregation of the mixture into highly ordered subdomains containing ligand 1a (green box) or 1b (red box). Additionally, several defects exhibiting coordination of neighboring bipyridine ligands of different lengths are highlighted (white boxes). Image size is 22 × 14 nm. (c) Schematic diagram of the reversible pyridyl–Fe bonding, the basis for active error correction by self-selection of ligands 1a and 1b into highly ordered subdomains. (d) Random packing would require breaking of one of the C–O Fe bonds and distortion of the other bond.
Fe bonds and distortion of the other bond.
Significantly improved error correction in the supramolecular Fe/1/2 system is achieved by reversing the principle selection control; i.e., the combination of components was changed to consist of two of the more labile bipyridine ligands 1a and 1b with only one bis-carboxylic acid ligand 2a. The Fe/1a/1b/2a mixture self-assembles into a supramolecular array of well ordered, rectangular compartments (Fig. 4b). A closer view reveals local spatial segregation into subdomains of the two distinctive heteroligand (1a × 2a and 1b × 2a) architectures as highlighted by the green and red rectangular frames, respectively. These subdomains within the 2D network can be considered as quasi-1D “ladder” rows, within which the two “side posts” consist of rows of carboxylate(2a)–Fe chains (horizontal in Fig. 4b) bridged with bipyridine ligand “rungs” of uniform size (either 1a or 1b in a given domain) at the Fe dimer nodes. The preferential binding of bipyridine ligands of uniform length within a ladder row indicates that the self-assembly of the mixture must involve both self-selection and self-recognition. With this mixture composition, the critical error correction process relies on the reversible coordination of the more labile bipyridine ligands 1a and 1b (Fig. 4c). The release of the pyridyl–Fe coordination in the case of negative self-recognition is favored compared with a distortion of the carboxylic–Fe dimer interaction (Fig. 4d). Thus, the read out in the self-assembly process occurs with higher degree of self-recognition, triggers spontaneous self-selection, and drives the self-assembly into locally segregated subdomains. This structural outcome of the assembly demonstrates that the self-selection is sufficiently strong to segregate ligands 1a and 1b, rather than form domains of higher entropy configurations with randomly distorted structures of Fe/1a/1b/2a (Fig. 4d). The efficient error correction of this structure is in sharp contrast to the previous mixture (Fig. 3), where the flexibility in assembly provides error tolerance leading to diminished structural order.
Besides defects due to the incomplete coverage of the surface (e.g., empty regions and linear bipyridine chains) (20) and network domain boundaries (vertical near the right edge of Fig. 4b), the presence of short bipyridine ligand 1a next to long bipyridine ligand 1b within a network domain is only rarely observed in the Fe/1a/1b/2a mixture (white frames in Fig. 4b). Because this coordination usually leads to singly bonded ligand species of 1, the detachment of the mismatched ligand and subsequent replacement with a new ligand of correct length should be favored. If a molecule of 1 has achieved a two-end bonding, the growth of the architecture will be stabilized by the gain of cooperativity, i.e., by the attachment of a larger number of ligands of the same length. In this sense, Fig. 4 can be interpreted as a “snap-shot” of an instructed mixture, which has already achieved segregation into small subdomains with a high level of internal organization and efficient local discrimination. The segregation is finally driven by the cooperativity of a coupled self-recognition/self-assembly process featuring efficient error-correction mechanisms.
Our investigations have demonstrated how error correction is improved by selection of components that provide (i) efficient reversibility of the Fe coordination bonds (pyridine–Fe versus carboxylate–Fe) and (ii) cooperative binding of the ligand components. Due to the restricted mobility of components at surfaces, efficient error-correction mechanisms, such as self-recognition and cooperatively amplified self-assembly, become of prime importance to achieve self-selection of the components. Surface-based studies allow (via STM) a unique “snap-shot” view on the evolution of multicomponent assemblies and can show with unprecedented resolution how supramolecular systems achieve high degrees of order, integration, and complexity by efficient self-selection. The multicomponent supramolecular self-assembly processes at surfaces can be considered as the 2D equivalent of the self-recognition of double helicate structures and other self-assembled molecular structures (21–24). The surface-assisted 2D self-assembly process is governed by similar thermodynamic rules and structural factors (e.g., interplay between read-out instruction of binding, conformational matching, and maximal occupation) as found in 3D supramolecular systems. In a broader perspective, insights into the formation of biologically active 3D compartments, representing the stage for the development of life at the organic/inorganic interface (25), can be obtained by such model studies of dynamic assembly, self-recognition, and self-selection. The unprecedented insight into the dynamics of model heteroassembly processes at the single-molecule level demonstrate critical assembly requirements for natural biological self-organization (25, 26).
Materials and Methods
The organic ligands were either purchased (ligands 2a and 2b) or synthesized (ligands 1a, 1b, and 2c) according to literature protocols (14, 27, 28). Evaporation and STM measurements were performed under ultrahigh vacuum conditions (∼2 × 10−10 mbar) using an atomically flat and clean Cu(100) surface prepared by repeated sputtering and annealing cycles. The substrate was held at ambient temperature while the ligands and Fe atoms were sequentially evaporated from Knudsen cells or an e-beam evaporator, and then was annealed to 450 K for 10 min. Prior work has demonstrated that such an annealing treatment efficiently deprotonates the carboxylic functional groups of the ligand family 2 to carboxylate groups (29, 30). The STM images were taken with a home-built microscope operated at room temperature in the constant current mode with a tunneling current of ≈0.1 nA and sample bias between 0.1 and 1.0 V.
Acknowledgments
This work was supported by European Commission Sixth Framework Program Project BIOMACH Grant NMP4-CT-2003-505-487 and by the European Science Foundation Project FunSMARTS II. S.L.T. was supported by the Alexander von Humboldt Foundation.
Abbreviation
- STM
scanning tunneling microscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Bonding energies given represent a first approximation based on calculations for carboxylate–Fe and pyridyl–Fe confined to a 2D plane in a vacuum (substrate influence neglected).
References
- 1.Lehn JM. Proc Natl Acad Sci USA. 2002;99:4763–4768. doi: 10.1073/pnas.072065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidel SR, Stang PJ. Acc Chem Res. 2002;35:972–983. doi: 10.1021/ar010142d. [DOI] [PubMed] [Google Scholar]
- 3.Caulder DL, Raymond KN. Acc Chem Res. 1999;32:975–982. [Google Scholar]
- 4.Caulder DL, Raymond KN. Dalton Trans. 1999:1185–1200. [Google Scholar]
- 5.Stang PJ, Olenyuk B. Acc Chem Res. 1997;30:502–518. [Google Scholar]
- 6.Spillmann H, Dmitriev A, Lin N, Messina P, Barth JV, Kern K. J Am Chem Soc. 2003;125:10725–10728. doi: 10.1021/ja0362353. [DOI] [PubMed] [Google Scholar]
- 7.Stepanow S, Lin N, Barth JV, Kern K. J Phys Chem B. 2006;110:23472–23477. doi: 10.1021/jp065066g. [DOI] [PubMed] [Google Scholar]
- 8.Lehn JM. Rep Prog Phys. 2004;67:249–265. [Google Scholar]
- 9.Ruben M, Payer D, Landa A, Comisso A, Gattinoni C, Lin N, Collin JP, Sauvage JP, De Vita A, Kern K. J Am Chem Soc. 2006;128:15644–15651. doi: 10.1021/ja063601k. [DOI] [PubMed] [Google Scholar]
- 10.Stohr M, Wahl M, Galka CH, Riehm T, Jung TA, Gade LH. Angew Chem Int Ed. 2005;44:7394–7398. doi: 10.1002/anie.200502316. [DOI] [PubMed] [Google Scholar]
- 11.Theobald JA, Oxtoby NS, Phillips MA, Champness NR, Beton PH. Nature. 2003;424:1029–1031. doi: 10.1038/nature01915. [DOI] [PubMed] [Google Scholar]
- 12.Barth JV, Costantini G, Kern K. Nature. 2005;437:671–679. doi: 10.1038/nature04166. [DOI] [PubMed] [Google Scholar]
- 13.Stepanow S, Lin N, Payer D, Schlickum U, Klappenberger F, Zoppellaro G, Ruben M, Brune H, Barth JV, Kern K. Angew Chem Int Ed. 2007;46:710–713. doi: 10.1002/anie.200603644. [DOI] [PubMed] [Google Scholar]
- 14.Stepanow S, Lingenfelder M, Dmitriev A, Spillmann H, Delvigne E, Lin N, Deng XB, Cai CZ, Barth JV, Kern K. Nature Mat. 2004;3:229–233. doi: 10.1038/nmat1088. [DOI] [PubMed] [Google Scholar]
- 15.Lin N, Stepanow S, Vidal F, Barth JV, Kern K. Chem Commun. 2005:1681–1683. doi: 10.1039/b418174c. [DOI] [PubMed] [Google Scholar]
- 16.Lehn JM. Supramolecular Chemistry Concepts and Perspectives. Weinheim, Germany: VHC; 1995. [Google Scholar]
- 17.Lin N, Stepanow S, Vidal F, Kern K, Alam MS, Stromsdorfer S, Dremov V, Muller P, Landa A, Ruben M. Dalton Trans. 2006:2794–2800. doi: 10.1039/b515728e. [DOI] [PubMed] [Google Scholar]
- 18.Ruben M. Angew Chem Int Ed. 2005;44:1594–1596. doi: 10.1002/anie.200462064. [DOI] [PubMed] [Google Scholar]
- 19.Seitsonen AP, Lingenfelder M, Spillmann H, Dmitriev A, Stepanow S, Lin N, Kern K, Barth JV. J Amer Chem Soc. 2006;128:5634–5635. doi: 10.1021/ja060180y. [DOI] [PubMed] [Google Scholar]
- 20.Tait SL, Langner A, Lin N, Stepanow S, Rajadurai C, Ruben M, Kern K. J Phys Chem C. 2007;111:10982–10987. [Google Scholar]
- 21.Baxter PNW, Lehn JM, Baum G, Fenske D. Chem Eur J. 1999;5:102–112. [Google Scholar]
- 22.Kramer R, Lehn JM, Marquis-Rigault A. Proc Natl Acad Sci USA. 1993;90:5394–5398. doi: 10.1073/pnas.90.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruben M, Rojo J, Romero-Salguero FJ, Uppadine LH, Lehn JM. Angew Chem Int Ed. 2004;43:3644–3662. doi: 10.1002/anie.200300636. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizawa M, Nakagawa J, Kurnazawa K, Nagao M, Kawano M, Ozeki T, Fujita M. Angew Chem Int Ed. 2005;44:1810–1813. doi: 10.1002/anie.200462171. [DOI] [PubMed] [Google Scholar]
- 25.Huber C, Wachtershauser G. Science. 1998;281:670–672. doi: 10.1126/science.281.5377.670. [DOI] [PubMed] [Google Scholar]
- 26.Dorr M, Kassbohrer J, Grunert R, Kreisel G, Brand WA, Werner RA, Geilmann H, Apfel C, Robl C, Weigand W. Angew Chem Int Ed. 2003;42:1540–1543. doi: 10.1002/anie.200250371. [DOI] [PubMed] [Google Scholar]
- 27.Biradha K, Hongo Y, Fujita M. Angew Chem Int Ed. 2000;39:3843–3845. doi: 10.1002/1521-3773(20001103)39:21<3843::AID-ANIE3843>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Fujita M, Oka H, Ogura K. Tetrahedr Lett. 1995;36:5247–5250. [Google Scholar]
- 29.Perry CC, Haq S, Frederick BG, Richardson NV. Surf Sci. 1998;409:512–520. [Google Scholar]
- 30.Stepanow S, Strunskus T, Lingenfelder M, Dmitriev A, Spillmann H, Lin N, Barth JV, Woll C, Kern K. J Phys Chem B. 2004;108:19392–19397. [Google Scholar]